5 Protein ID conversion
If you have handled proteomics or metabolomics data (e.g. mass spectrometry), you may need to convert protein to gene id before downstream analysis.
5.1 Example data
For example, in this data article Dataset from proteomic analysis of rat, mouse, and human liver microsomes and S9 fractions , we could get rat proteomic analysis data
Firstly, take a look at the data:
colnames(rat_prodata)## [1] "GI" "Acc" "Gene" "Name" "GRAVY" "Length" "Sequence"head(rat_prodata, 5)## GI Acc Gene
## 1 112774 P27364 3BHS5_RAT
## 2 112889 P17475 A1AT_RAT
## 3 112893 P14046 A1I3_RAT
## 4 112987 P00507 AATM_RAT
## 5 113016 P15650 ACADL_RAT
## Name GRAVY Length
## 1 3 beta-hydroxysteroid dehydrogenase type 5 -0.2168901 373
## 2 Alpha-1-antiproteinase -0.1576642 411
## 3 Alpha-1-inhibitor 3 -0.2164523 1477
## 4 Aspartate aminotransferase, mitochondrial -0.2295349 430
## 5 Long-chain specific acyl-CoA dehydrogenase, mitochondrial -0.2227907 430
## Sequence
## 1 MPGWSCLVTGAGGFLGQRIVQMLVQEKELQEVRVLYRTFSPKHKEELSKLQTKAKVTVLRGDIVDAQFLRRACQGMSVIIHTAAALDIAGFLPRQTILDVNVKGTQLLLDACVEASVPAFIYSSSTGVAGPNSYKETILNDREEEHRESTWSNPYPYSKRMAEKAVLAANGSILKNGGTFHTCALRLPFIYGEESQIISTMVNRALKNNSIIKRHATFSIANPVYVGNAAWAHILAARGLRDPEKSQSIQGQFYYISDDTPHQSYDDLNYTLSKEWGFCLDSSWSLPLPLLYWLAFLLETVSFLLRPFYNYRPPFNRFMVTILNSVFTISYKKAQRDLGYEPLVSWEEAKQKTSEWIGTLVEQHRETLDTKSQ
## 2 MAPSISRGLLLLAALCCLAPSFLAEDAQETDTSQQDQSPTYRKISSNLADFAFSLYRELVHQSNTSNIFFSPMSITTAFAMLSLGSKGDTRKQILEGLEFNLTQIPEADIHKAFHHLLQTLNRPDSELQLNTGNGLFVNKNLKLVEKFLEEVKNNYHSEAFSVNFADSEEAKKVINDYVEKGTQGKIVDLMKQLDEDTVFALVNYIFFKGKWKRPFNPEHTRDADFHVDKSTTVKVPMMNRLGMFDMHYCSTLSSWVLMMDYLGNATAIFLLPDDGKMQHLEQTLTKDLISRFLLNRQTRSAILYFPKLSISGTYNLKTLLSSLGITRVFNNDADLSGITEDAPLKLSQAVHKAVLTLDERGTEAAGATVVEAVPMSLPPQVKFDHPFIFMIVESETQSPLFVGKVIDPTR
## 3 MKKDREAQLCLFSALLAFLPFASLLNGNSKYMVLVPSQLYTETPEKICLHLYHLNETVTVTASLISQRGTRKLFDELVVDKDLFHCVSFTIPRLPSSEEEESLDINIEGAKHKFSERRVVLVKNKESVVFVQTDKPMYKPGQSVKFRVVSMDKNLHPLNELFPLAYIEDPKMNRIMQWQDVKTENGLKQLSFSLSAEPIQGPYKIVILKQSGVKEEHSFTVMEFVLPRFGVDVKVPNAISVYDEIINVTACATYTYGKPVPGHVKISLCHGNPTFSSETKSGCKEEDSRLDNNGCSTQEVNITEFQLKENYLKMHQAFHVNATVTEEGTGSEFSGSGRIEVERTRNKFLFLKADSHFRHGIPFFVKVRLVDIKGDPIPNEQVLIKARDAGYTNATTTDQHGLAKFSIDTNGISDYSLNIKVYHKEESSCIHSSCTAERHAEAHHTAYAVYSLSKSYIYLDTEAGVLPCNQIHTVQAHFILKGQVLGVLQQIVFHYLVMAQGSILQTGNHTHQVEPGESQVQGNFALEIPVEFSMVPVAKMLIYTILPDGEVIADSVKFQVEKCLRNKVHLSFSPSQSLPASQTHMRVTASPQSLCGLRAVDQSVLLQKPEAELSPSLIYDLPGMQDSNFIASSNDPFEDEDYCLMYQPIAREKDVYRYVRETGLMAFTNLKIKLPTYCNTDYDMVPLAVPAVALDSSTDRGMYESLPVVAVKSPLPQEPPRKDPPPKDPVIETIRNYFPETWIWDLVTVNSSGVTELEMTVPDTITEWKAGALCLSNDTGLGLSSVASFQAFQPFFVELTMPYSVIRGEAFTLKATVLNYLPTSLPMAVLLEASPDFTAVPVENNQDSYCLGANGRHTSSWLVTPKSLGNVNFSVSAEARQSPGPCGSEVATVPETGRKDTVVKVLIVEPEGIKKEHTFSSLLCASDAELSETLSLLLPPTVVKDSARAHFSVMGDILSSAIKNTQNLIQMPYGCGEQNMVLFAPNIYVLKYLNETQQLTEKIKSKALGYLRAGYQRELNYKHKDGSYSAFGDHNGQGQGNTWLTAFVLKSFAQARAFIFIDESHITDAFTWLSKQQKDSGCFRSSGSLLNNAMKGGVDDEITLSAYITMALLESSLPDTDPVVSKALSCLESSWENIEQGGNGSFVYTKALMAYAFALAGNQEKRNEILKSLDKEAIKEDNSIHWERPQKPTKSEGYLYTPQASSAEVEMSAYVVLARLTAQPAPSPEDLALSMGTIKWLTKQQNSYGGFSSTQDTVVALDALSKYGAATFSKSQKTPSVTVQSSGSFSQKFQVDKSNRLLLQQVSLPYIPGNYTVSVSGEGCVYAQTTLRYNVPLEKQQPAFALKVQTVPLTCNNPKGQNSFQISLEISYMGSRPASNMVIADVKMLSGFIPLKPTVKKLERLGHVSRTEVTTNNVLLYLDQVTNQTLSFSFIIQQDIPVKNLQPAIVKVYDYYETDEVAFAEYSSPCSSDDQNV
## 4 MALLHSGRVLSGMAAAFHPGLAAAASARASSWWTHVEMGPPDPILGVTEAFKRDTNSKKMNLGVGAYRDDNGKPYVLPSVRKAEAQIAGKNLDKEYLPIGGLADFCKASAELALGENSEVLKSGRFVTVQTISGTGALRVGASFLQRFFKFSRDVFLPKPSWGNHTPIFRDAGMQLQGYRYYDPKTCGFDFSGALEDISKIPEQSVLLLHACAHNPTGVDPRPEQWKEMAAVVKKKNLFAFFDMAYQGFASGDGDKDAWAVRHFIEQGINVCLCQSYAKNMGLYGERVGAFTVVCKDAEEAKRVESQLKILIRPLYSNPPLNGARIAATILTSPDLRKQWLQEVKGMADRIISMRTQLVSNLKKEGSSHNWQHITDQIGMFCFTGLKPEQVERLTKEFSVYMTKDGRISVAGVTSGNVGYLAHAIHQVTK
## 5 MAARLLLRSLRVLSARSATLPPPSARCSHSGAEARLETPSAKKLTDIGIRRIFSSEHDIFRESVRKFFQEEVIPYHEEWEKAGEVSRELWEKAGKQGLLGINIAEKHGGIGGDLLSTAVTWEEQAYSNCTGPGFSLHSDIVMPYIANYGTKEQIEQFIPQMTAGKCIGAIAMTEPGAGSDLQGVRTNAKRSGSDWILNGSKVFITNGWLSDLVIVVAVTNREARSPAHGISLFLVENGMKGFIKGKKLHKMGMKAQDTAELFFEDVRLPASALLGEENKGFYYLMQELPQERLLIADLAISACEFMFEETRNYVRQRKAFGKTVAHIQTVQHKLAELKTNICVTRAFVDSCLQLHETKRLDSASASMAKYWASELQNTVAYQCVQLHGGWGYMWEYPIAKAYVDARVQPIYGGTNEIMKELIARQIVSDSThe second column is rat protein id, and the third one is gene name.
5.2 Id conversion
Here we will convert protein id to gene symbol and compare with the original one.
protein_id <- rat_prodata[, "Acc"]
gname <- transId(protein_id, "symbol", "rat", unique = T)
new_res <- merge(rat_prodata, gname,
by.x = "Acc", by.y = "input_id", all.x = T
) %>%
dplyr::relocate(symbol, .after = Gene)
head(new_res, 5)## Acc GI Gene symbol
## 1 A0FKI7 123797828 ACBD5_RAT Acbd5
## 2 A0JPQ8 158512263 ALKMO_RAT Agmo
## 3 A0JPQ9 134035401 CHID1_RAT LOC100911881
## 4 A1L134 221222583 AUP1_RAT Aup1
## 5 A1L1J9 171704593 LMF2_RAT Lmf2
## Name GRAVY Length
## 1 Acyl-CoA-binding domain-containing protein 5 -0.81245059 506
## 2 Alkylglycerol monooxygenase 0.23579418 447
## 3 Chitinase domain-containing protein 1 -0.20229008 393
## 4 Ancient ubiquitous protein 1 -0.09902439 410
## 5 Lipase maturation factor 2 0.04757835 702
## Sequence
## 1 MLFLSFYAGSWESWICCCCVIPVDRPWDRGRRWQLEMADTRSVYETRFEAAVKVIQSLPKNGSFQPTNEMMLRFYSFYKQATEGPCKLSRPGFWDPIGRYKWDAWSSLGDMTKEEAMIAYVEEMKKIIETMPMTEKVEELLHVIGPFYEIVEDKKNSKSSDLTSDLGNVLTSSNAKAVNGKAESSDSGAESEEEEAQEELKGAEQSGSDDKKMMTKSTDKNLEIIVTNGYKDSFAQDSDIHTDSSRSARRSEDKKPTDQSSQQTGNTVLCVHQDTNEDPGEDASGVHHLTSDSDSEVYCDSMEQFGQEEYYLGGDPAQHLEGSGFCEDAQLSPGNGSIGKMQMRAVKGKGEVKHGGEDGRSSSGTPHREKRGGESEDISGVRRGRGHRMPHLSEGTKGRQVGSGGDGERWGSDRGSRGSLNEQIALVLIRLQEDMQNVLQRLHKLETLTASQAKLSWQTSNQPSSQRPSWWPFEMSPGALAFAIIWPFIAQWLVHLYYQRRRRKLN
## 2 MRNPGAQDNVSVSQGMRAMFYMMKPSETAFQTVEEVPDYVKKATPFFIFLILLELVVSWILKGKPSGRLDDILTSMSAGVVSRLPNLFFRSLEVTSYIYIWENYRVCELPWDSPWTWYLTFLGVDFGYYWFHRMAHEINIIWAAHQAHHSSEDYNLSTALRQSVLQQYSSWVFYCPLALFVPPSVFAVHIQFNLLYQFWIHTEVIRTLGPLELVLNTPSHHRVHHGRNRYCIDKNYAGTLIIWDRIFGTFEAENEQVIYGLTHPIGTFEPFKVQFHHLLYIWTTFWATPGFCHKFSVLFKGPGWGPGKPRLGLSEEIPEVTGQEVPFTSSASQFLKIYAVLQFAVMLVFYEETFANTAVLSQVTILLRICFIILTLTSIGFLLDQRPKAAIVETLRCLLFLTLYRFGHLKPLIESLSFAFEIFFSVCIAFWGVRSITHLASGSWKKP
## 3 MWPLLHVLWLALVCGSVHTTLSKSDAKKAASKTLLEKTQFSDKPVQDRGLVVTDIKAEDVVLEHRSYCSARARERNFAGEVLGYVTPWNSHGYDVAKVFGSKFTQISPVWLQLKRRGREMFEITGLHDVDQGWMRAVKKHAKGVRIVPRLLFEDWTYDDFRSVLDSEDEIEELSKTVVQVAKNQHFDGFVVEVWSQLLSQKHVGLIHMLTHLAEALHQARLLVILVIPPAVTPGTDQLGMFTHKEFEQLAPILDGFSLMTYDYSTSQQPGPNAPLSWIRACVQVLDPKSQWRSKILLGLNFYGMDYAASKDAREPVIGARYIQTLKDHRPRVVWDSQAAEHFFEYKKNRGGRHVVFYPTLKSLQVRLELARELGVGVSIWELGQGLDYFYDLL
## 4 MEPPPAPGPERLFDSHRLPSDGFLLLALLLYAPVGLCLLVLRLFLGLHVFLVSCALPDSVLRRFVVRTMCAVLGLVARQEDSGLRDHRVRVLISNHVTPFDHNIVNLLTTCSTPLLNSPPSFVCWSRGFMEMDRRVELVESLKKFCASTRLPPTPLLLFPEEEATNGREGLLRFSSWPFSIQDVVQPLTLQVQRPLVSVTVSDASWVSELLWSLFVPFTVYQVRWLHPIRRQLGEENEEFALRVQQLVAKELGQIGTRLTPADKAEHMKRQRHPRLRPQSVQSSFPSPPSPSSDVQLTILAQRVKEVLPHVPLNVIQRDLARTGCVDLTITNLLEGAVAFMPEDVTEGSQSLPTASAPKFPSSGLVTPQPTALTFAKSSWARQESLQERKQALYEYARRRFRERQAQEAE
## 5 MASARVPQQLFLQGVAAVYLFAFASLYTQIPGLYGPEGILPARRTLRPQGKGRWQQLWETPTILWEAPRLGLDTAQGLDLLTLLGTVLALGALLLNSLRHPFIYLLLWAAYLSACQVGQVFLYFQWDSLLLETGFLAILVAPLRRPSKHKIPQGGLAGALPHEDLPFWLVRWLLFRLMFASGVVKLTSRCPAWWGLTALTYHYETQCLPTPAAWFAHHLPVWLHRLSVVATFLIEIAVPPLFFAPIRRLRLSAFYAQALLQILIIITGNYNFFNLLTLVLTTALLDDRHLSAEPELRCHKKMPTSWPKTLLTSLSLMLELTVYGLLAYGTIYYFGLEVDWQQQIVLSKTTFTFHQFSQWLKMVTLPTVWLGTASLAWELLIALWRWIQVQGWSRKFFAGIQLSVLGTATVFLFLISLVPYSYVEPGTHGRLWTGAHRLFSSVEHLQLANSYGLFRRMTGLGGRPEVVLEGSHDGHHWTEIEFMYKPGNVSRPPPFLIPHQPRLDWQMWFAALGPHTHSPWFTSLVLRLLQGKEPVIRLIQNQVANYPFREQPPTYLRAQRYKYWFSKPGDQSRWWHRQWVEEFFPSVSLGDPTLETLLQQFGLKDKSPPRARSSKNALAQTLNWVRAQLSPLEPSILLWGLLGAVVAIRVVRTLLTPRPLQSSKQTREEKRKQAPKKDSRAVSEQTAPNSNSNGSWAPRRKKMaybe you have noticed that some gene names are different from the original data. For example, the second one: A0JPQ8 is called Alkmo before, but the latest name is Agmo.
Some proteins have new gene names like above, while some may have no names for now.
old_name <- unique(new_res$Gene) %>%
stringr::str_remove_all("_RAT")
new_name <- unique(toupper(new_res$symbol))
new_res[which(is.na(new_res$symbol)), ] %>%
dplyr::select(Acc, Gene, symbol)## Acc Gene symbol
## 103 P01835 KACB_RAT <NA>
## 215 P0C169 H2A1C_RAT <NA>
## 305 P16391 HA12_RAT <NA>
## 485 P50169 RDH3_RAT <NA>
## 544 P62161 CALM_RAT <NA>
## 620 P81828 UP2_RAT <NA>
## 623 P83121 UP3_RAT <NA>
## 950 Q6P6G2 CF089_RAT <NA>We could see the result above: the protein id P01835 has replaced the gene name with NA.
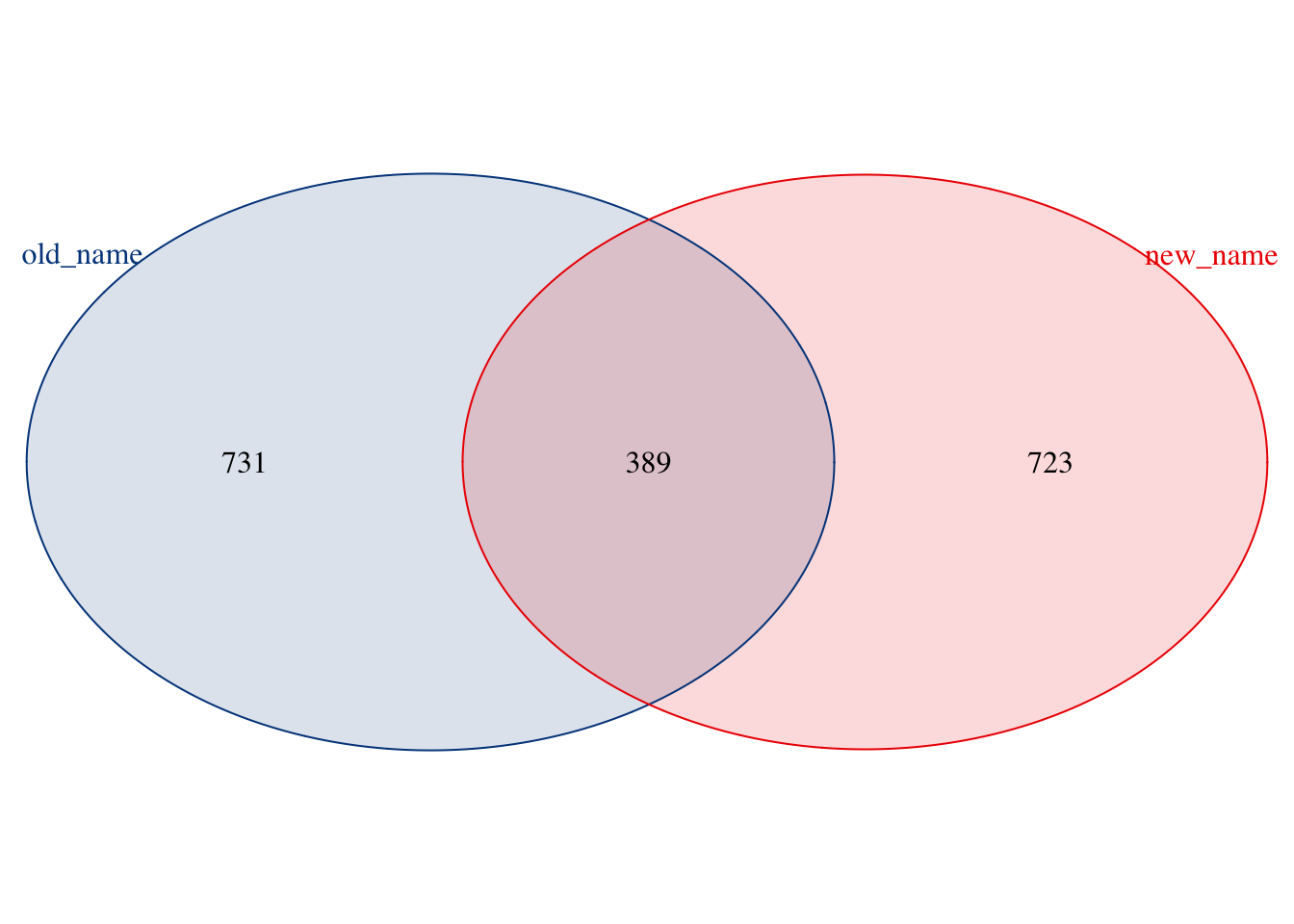
The Venn plot shows that primary protein ids have changed their gene names. If users continue to use the old gene names, they may lose the latest gene information even enriched pathways.