4 Gene ID conversion
If you have handled genomics analysis, then you may spend much time converting gene identifiers to other types.
For example, gene set enrichment analysis is very common in omics downstream process which is mainly based on MsigDb. According to its release note, since MSigDB 7.0, identifiers for genes are mapped to their HGNC approved Gene Symbol and NCBI Gene ID through annotations extracted from Ensembl’s BioMart data service, and will be updated at each MSigDB release with the latest available version of Ensembl. Currently, MSigDB 7.5 has updated human gene annotations to Ensembl v105.
Hence, id conversion could help keeping update with the latest annotations.
The most common gene id types are symbol (e.g. TP53),NCBI Entrez (e.g. 7157) and Ensembl (e.g. ENSG00000141510). ID conversion among different types is very common and there are two main concerns: performance and flexibility.
4.1 Current tools
Currently various tools are available, however different methods has their drawbacks:
- biomaRt: The R package is frequently used to access up-to-date Ensembl database. It has to access online database each time and built-in parameters are too many.
- clusterProfiler: The R package depends on Bioconductor annotation packages to extract NCBI gene. NCBI database updates every day while Bioconductor package updates nearly every six months. Besides, it only supports 20 organisms and user should download and load annotation data before mapping.
- gProfiler: The web server supports 40 types of IDs for more than 60 species. But it only supports one-to-one conversion, for example, if user want to convert gene symbol to both Entrez and Ensembl, it could not happen. Besides, the output result is not convenient to further process.
- DAVID: The web server interface is not user-friendly.
- UniProt: The large protein database may support almost all species but user needs to specify input id type and only converts to protein id.
- GIDcon: The web server only covers human, mouse and rat. User must seperates gene ids with comma if does batch query. Five symbols will take 8 secs to convert.
Considered above problems, genekitr combines both Ensembl and NCBI database to provide more comprehensive result while optimizes running speed.
Also it supports 195 vertebrate species, 120 plant species and 2 bacteria species (see also session 1.1).
4.2 Basic usage
transId provides five arguments:
id: gene id (symbol, Entrez or Ensembl), protein id (see also session 4.2) or microarray id (see also session 5.1)transTo: the conversion target type. User could select more than one from “symbol”, “entrez”, “ensembl” or “uniprot”. Besides, abbreviation is acceptable (e.g. “ens” is fine for “ensembl” but “en” will raise error because it could not separate from “entrez”)org: organism name, default is humanunique:TRUEorFALSE(default). IfTRUE, only returns one matched id.keepNA:TRUEorFALSE(default). If some id has no matched one, keep it or not.
## input_id ensembl
## 1 AKT3 ENSG00000275199
## 2 AKT3 ENSG00000117020
## 3 SSX6P ENSG00000171483
## 4 BCC7 ENSG00000141510# fruit fly genes
transId(
id = c("10178780", "10178777", "10178786", "10178792"),
transTo = "sym", org = "fly"
)## input_id symbol
## 1 10178780 CR33929
## 2 10178777 CG42835
## 3 10178786 Su(Ste):CR40820
## 4 10178792 CG42694See the human example above, due to NA was removed, the result order is slightly different from input id.
If you want to get the same order, you can set keepNA = TRUE:
transId(id, transTo = "ens", keepNA = TRUE)## input_id ensembl
## 1 AKT3 ENSG00000275199
## 2 AKT3 ENSG00000117020
## 3 SSX6P ENSG00000171483
## 4 FAKE_ID <NA>
## 5 BCC7 ENSG00000141510If you want to get one-to-one mapping, you can set unique = TRUE and the result with more comprehensive information will return:
transId(id, transTo = "ens", keepNA = TRUE, unique = TRUE)## input_id ensembl
## 21 AKT3 ENSG00000117020
## 1 SSX6P ENSG00000171483
## 2 FAKE_ID <NA>
## 3 BCC7 ENSG000001415104.3 Features
4.3.1 f1: convert to several types
transTo could accepts more than one type:
## input_id ensembl entrezid
## 1 AKT3 ENSG00000275199 10000
## 2 AKT3 ENSG00000117020 10000
## 3 SSX6P ENSG00000171483 280657
## 4 BCC7 ENSG00000141510 71574.3.2 f2: distinguish from symbol and alias
One big common problem for current tools is confusion of gene symbol and gene alias.
Gene alias is an alternative name of official gene symbol, like “BCC7” is actually alias for “TP53”. Sometimes gene alias is more widely used than official symbol. For example, “PDL1” is the alias for “CD274” in fact.
Due to combination of NCBI and Ensembl metadata, transId could easily recognize gene symbol and alias by setting transTo = 'symbol'.
## input_id symbol
## 1 BCC7 TP53
## 2 TP53 TP53
## 3 PD1 PDCD1
## 4 PD1 SNCA
## 5 PD1 SPATA2
## 6 PDL1 CD274
## 7 TET2 TET24.3.3 f3: no worry for Ensembl version
An Ensembl stable ID consists of five parts: ENS(species)(object type)(identifier). (version)
For example, ENSG00000141510.11 is TP53 while the version .11 may confuse many tools.
However using transId, user will not worry this issue any more.
transId("ENSG00000141510.11", "symbol")## input_id symbol
## 1 ENSG00000141510 TP534.4 Excercise 1: DEG result
The built-in example data is differentially expressed genes from: GSE42872
##
## down stable up
## 451 17009 579id <- deg$entrezid
length(id)## [1] 18039
4.4.1 clusterProfiler::bitr
User needs to specify input id type and converted type from columns(org.Hs.eg.db).
# User must firstly install the org.Hs.eg.db
# BiocManager::install('org.Hs.eg.db')
library(org.Hs.eg.db)
library(clusterProfiler)
AnnotationDbi::columns(org.Hs.eg.db)## [1] "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT" "ENSEMBLTRANS"
## [6] "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "GENENAME"
## [11] "GO" "GOALL" "IPI" "MAP" "OMIM"
## [16] "ONTOLOGY" "ONTOLOGYALL" "PATH" "PFAM" "PMID"
## [21] "PROSITE" "REFSEQ" "SYMBOL" "UCSCKG" "UNIGENE"
## [26] "UNIPROT"bitr_res <- bitr(id, fromType = "ENTREZID", toType = "SYMBOL", OrgDb = "org.Hs.eg.db")
bitr_sym <- unique(bitr_res$SYMBOL)
length(bitr_sym)## [1] 18036
4.4.2 biomaRt::getBM
The initializing process depends mainly on Ensembl’s web access speed. Usually, it may take tens of seconds.
# initializing...
ensembl <- biomaRt::useEnsembl(
biomart = "genes",
dataset = "hsapiens_gene_ensembl"
)
# conversion
biomart_res <- biomaRt::getBM(
attributes = c("entrezgene_id", "external_gene_name"),
filters = c("entrezgene_id"),
mart = ensembl,
values = id
)biomart_sym <- unique(biomart_res$external_gene_name) %>%
stringi::stri_remove_empty()
length(biomart_sym)## [1] 16085
4.4.3 genekitr::transId
transid_res <- genekitr::transId(id, transTo = "sym", unique = T)
transid_sym <- unique(transid_res$symbol)
length(transid_sym)## [1] 180394.4.4 compare three results
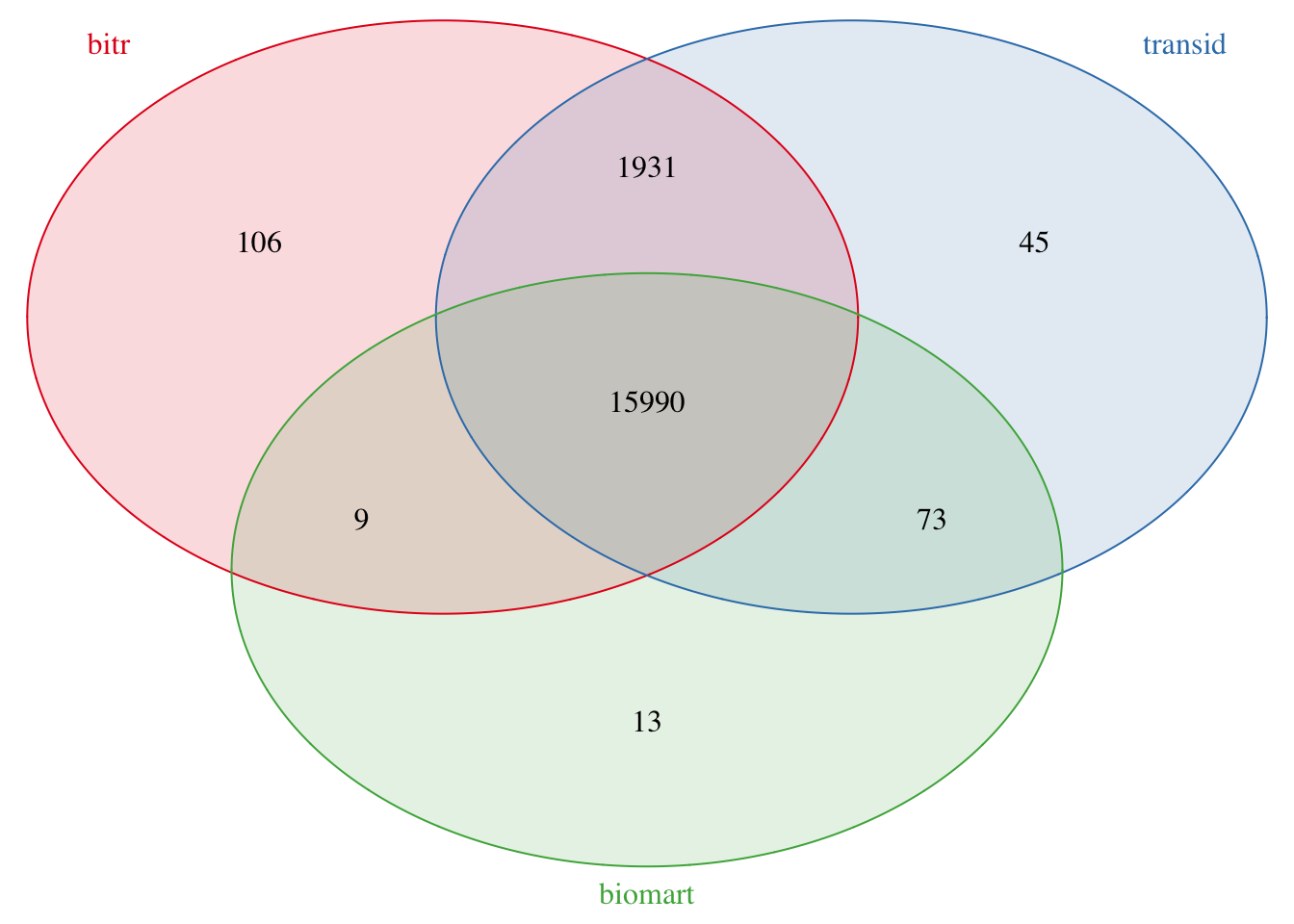
4.4.4.1 bitr vs transId
There are 117 genes only in bitr result:
## [1] "C19orf71" "LINC00476" "LOC100129484" "PP2672" "LOC100130449"Let’s take a look for the first one LINC00337, which is converted from gene 148645
## [1] "148645"When we search it on NCBI and we could see the official symbol is “ICMT-DT” now while “LINC00337” is an alias. When we look back into transId result, it keeps update with NCBI web.
## [1] "ICMT-DT"Besides, there are 118 unique symbols in transId result:
## [1] "TEKTIP1" "ERCC6L2-AS1" "LINC02983" "LINC02961" "GPC1-AS1"The above gene ICMT-DT is listed.
4.4.4.2 biomart vs transId
There are 13 genes only in biomart result:
## [1] "KLRK1" "FLJ37453" "C11orf49" "C19orf71" "ZNF761" "LINC01588"## [1] 100528032When we search NCBI 100528032 and we could see the official symbol is “KLRC4-KLRK1”. When we look back into transId and bitr result, they are the same with NCBI web.
## [1] "KLRC4-KLRK1"## [1] "KLRC4-KLRK1"4.5 Excercise 2: mixture of gene symbol and alias
Compared with Entrez and Ensembl, gene symbol and alias is hard to distinguish. In real science life, the impact of misunderstanding both names is huge.
Takehara et al. (2018) had to retract the published paper by its authors just because they misused “TAZ” as the actual research target “WWTR1” in which “TAZ” is an alias of “WWTR1”.
The similar issues have published in paper: The risks of using unapproved gene symbols. Of course, it is not only happens in human research. So we must raise alert of gene alias.
4.5.1 HGNChelper vs transId
HGNChelper is mainly built for human research, so here we will give simple human gene list as example.
id <- c("TBET", "B220", "BCC7", "PD1", "PDL1")
transid_res <- genekitr::transId(id, transTo = "sym", unique = FALSE)
transid_res## input_id symbol
## 1 TBET TBX21
## 2 B220 PTPRC
## 3 BCC7 TP53
## 4 PD1 PDCD1
## 5 PD1 SNCA
## 6 PD1 SPATA2
## 7 PDL1 CD274hgnc_res <- HGNChelper::checkGeneSymbols(id)
hgnc_res## x Approved Suggested.Symbol
## 1 TBET FALSE <NA>
## 2 B220 FALSE <NA>
## 3 BCC7 FALSE <NA>
## 4 PD1 FALSE PDCD1 /// SNCA /// SPATA2
## 5 PDL1 FALSE CD274The first three genes are omitted in HGNChelper.
4.5.2 GeneSymbolThesarus vs transId
GeneSymbolThesarus is a function of Seurat which finds current gene symbols from the HUGO Gene Nomenclature Committee (HGNC). It will search online so the speed will not fast.
## Attaching SeuratObject##
## Attaching package: 'Seurat'## The following object is masked from 'package:DT':
##
## JSstart <- Sys.time()
seurat_res <- Seurat::GeneSymbolThesarus(symbols = id)
end <- Sys.time()
(end - start)## Time difference of 57.88858 secsseurat_res## PDL1
## "CD274"Seurat only returns one records.
4.5.3 alias2Symbol vs transId
alias2Symbol is a function of limma which depends on organism annotation package of Bioconductor.
limma_res <- limma::alias2Symbol(id, species = "Hs")
# compare with transId result
id## [1] "TBET" "B220" "BCC7" "PD1" "PDL1"limma_res## [1] "CD274" "TBX21" "PDCD1" "PTPRC" "SNCA" "TP53" "SPATA2"transid_res$symbol## [1] "TBX21" "PTPRC" "TP53" "PDCD1" "SNCA" "SPATA2" "CD274"It seems that alias2Symbol result remains disorder from input id, which may cause misunderstanding for users, while transId keeps the original order (if user want to get one-to-one match, just pass unique = TRUE to function).