8 ORA analysis
As the paper “Urgent need for consistent standards in functional enrichment analysis” mentioned, popular functional enrichment tools can be classified into two main categories: (1) over-representation analysis (ORA) and (2) functional class scoring (FCS) .
Knidly Note: No matter which method you choose, the first thing is always the preparation of gene sets.
8.1 ORA method introduction
According to paper “Over-representation of correlation analysis (ORCA): a method for identifying associations between variable sets”:
Over-representation analysis (ORA) is a simple method for objectively deciding whether a set of variables of known or suspected biological relevance, such as a gene set or pathway, is more prevalent in a set of variables of interest than we expect by chance.
ORA usually has these procedures:
- Identify features, such as mRNA, proteins or microarray probes, considered “differentially expressed” in two or more conditions
- Determine the number of differentially expressed features in each pathway
- For each pathway, using a hypergeometric distribution to calculate a probability value ( P -value) of obtaining the number of differentially expressed features against the background list of all features
8.2 Basic usage
The simplest arguments are:
-
id: gene id of Entrez (numeric id is also supported), Ensembl, Symbol (alias is also accepted) or Uniprot -
geneset: gene set is a two-column data frame with term id and gene id. (It’s recommended to use geneset) -
p_cutoff: numeric of cutoff for both pvalue and adjusted pvalue, default is 0.05. -
q_cutoff: numeric of cutoff for qvalue, default is 0.15.
8.2.1 1st step: prepare input IDs
To show the various input ID types, we choose Entrez ID, Symbol from human and Uniprot from rat protein example data.
data(geneList, package = "genekitr")
entrez_id <- names(geneList)[abs(geneList) > 2]
head(entrez_id, 5)## [1] "948" "1638" "158471" "10610" "6947"symbol_id <- c("TP53","BRCA1","TET2") # all official symbols
mix_symbol_id <- c("BCC7","BRCA1","TET2") # mixture of official symbol and alias
unip_id <- rat_prodata$Acc[1:100]
head(unip_id, 5)## [1] "P27364" "P17475" "P14046" "P00507" "P15650"8.2.2 2nd step: prepare gene set
For more details, please refer to chapter7
Here we prepare GO MF gene set:
8.2.3 3rd step: ORA analysis
For non-symbol genes (e.g. Entrez IDs):
## Hs_MF_ID Description
## GO:0000217 GO:0000217 DNA secondary structure binding
## GO:0005041 GO:0005041 low-density lipoprotein particle receptor activity
## GO:0030228 GO:0030228 lipoprotein particle receptor activity
## GeneRatio BgRatio pvalue p.adjust qvalue
## GO:0000217 0.032 36/18543 9.808774e-05 0.02180140 0.01984400
## GO:0005041 0.024 15/18543 1.282436e-04 0.02180140 0.01984400
## GO:0030228 0.024 18/18543 2.266171e-04 0.02568327 0.02337734
## geneID geneID_symbol Count FoldEnrich
## GO:0000217 4208/641/7516/63967 MEF2C/BLM/XRCC2/CLSPN 4 16.48267
## GO:0005041 948/53353/3949 CD36/LRP1B/LDLR 3 29.66880
## GO:0030228 948/53353/3949 CD36/LRP1B/LDLR 3 24.72400
## RichFactor
## GO:0000217 0.1111111
## GO:0005041 0.2000000
## GO:0030228 0.1666667For symbol genes without alias:
## Hs_MF_ID Description
## GO:0031625 GO:0031625 ubiquitin protein ligase binding
## GO:0044389 GO:0044389 ubiquitin-like protein ligase binding
## GO:0001094 GO:0001094 TFIID-class transcription factor complex binding
## GO:0097371 GO:0097371 MDM2/MDM4 family protein binding
## GO:0140666 GO:0140666 annealing activity
## GO:0035035 GO:0035035 histone acetyltransferase binding
## GeneRatio BgRatio pvalue p.adjust qvalue geneID
## GO:0031625 0.6666667 301/18543 0.0007794317 0.01891673 0.004424966 TP53/BRCA1
## GO:0044389 0.6666667 320/18543 0.0008805032 0.01891673 0.004424966 TP53/BRCA1
## GO:0001094 0.3333333 10/18543 0.0016170760 0.01891673 0.004424966 TP53
## GO:0097371 0.3333333 11/18543 0.0017786877 0.01891673 0.004424966 TP53
## GO:0140666 0.3333333 13/18543 0.0021018587 0.01891673 0.004424966 TP53
## GO:0035035 0.3333333 23/18543 0.0037166674 0.02362933 0.005527329 TP53
## Count FoldEnrich RichFactor
## GO:0031625 2 41.06977 0.006644518
## GO:0044389 2 38.63125 0.006250000
## GO:0001094 1 618.10000 0.100000000
## GO:0097371 1 561.90909 0.090909091
## GO:0140666 1 475.46154 0.076923077
## GO:0035035 1 268.73913 0.043478261For symbol genes with alias:
## Hs_MF_ID Description
## GO:0031625 GO:0031625 ubiquitin protein ligase binding
## GO:0044389 GO:0044389 ubiquitin-like protein ligase binding
## GO:0001094 GO:0001094 TFIID-class transcription factor complex binding
## GO:0097371 GO:0097371 MDM2/MDM4 family protein binding
## GO:0140666 GO:0140666 annealing activity
## GO:0035035 GO:0035035 histone acetyltransferase binding
## GeneRatio BgRatio pvalue p.adjust qvalue geneID
## GO:0031625 0.6666667 301/18543 0.0007794317 0.01891673 0.004424966 BCC7/BRCA1
## GO:0044389 0.6666667 320/18543 0.0008805032 0.01891673 0.004424966 BCC7/BRCA1
## GO:0001094 0.3333333 10/18543 0.0016170760 0.01891673 0.004424966 BCC7
## GO:0097371 0.3333333 11/18543 0.0017786877 0.01891673 0.004424966 BCC7
## GO:0140666 0.3333333 13/18543 0.0021018587 0.01891673 0.004424966 BCC7
## GO:0035035 0.3333333 23/18543 0.0037166674 0.02362933 0.005527329 BCC7
## geneID_symbol Count FoldEnrich RichFactor
## GO:0031625 TP53/BRCA1 2 41.06977 0.006644518
## GO:0044389 TP53/BRCA1 2 38.63125 0.006250000
## GO:0001094 TP53 1 618.10000 0.100000000
## GO:0097371 TP53 1 561.90909 0.090909091
## GO:0140666 TP53 1 475.46154 0.076923077
## GO:0035035 TP53 1 268.73913 0.043478261For Uniprot proteins:
## Rn_MF_ID Description GeneRatio BgRatio
## GO:0004497 GO:0004497 monooxygenase activity 0.20000000 165/17190
## GO:0008395 GO:0008395 steroid hydroxylase activity 0.14736842 64/17190
## GO:0020037 GO:0020037 heme binding 0.18947368 179/17190
## GO:0046906 GO:0046906 tetrapyrrole binding 0.18947368 188/17190
## GO:0005506 GO:0005506 iron ion binding 0.16842105 208/17190
## GO:0070330 GO:0070330 aromatase activity 0.09473684 41/17190
## pvalue p.adjust qvalue
## GO:0004497 3.836117e-20 1.112474e-17 7.954895e-18
## GO:0008395 3.494524e-19 5.067060e-17 3.623270e-17
## GO:0020037 4.824632e-18 4.663811e-16 3.334921e-16
## GO:0046906 1.172685e-17 8.501966e-16 6.079446e-16
## GO:0005506 2.813821e-14 1.632016e-12 1.166995e-12
## GO:0070330 9.884697e-13 4.777603e-11 3.416290e-11
## geneID
## GO:0004497 P24483/P17178/P04800/P20816/P18125/P15149/P00176/P13107/P05178/P05179/P08683/P11510/P20814/P19225/P10633/P10634/P12939/P11715/P23457
## GO:0008395 P24483/P17178/P04800/P18125/P15149/P00176/P13107/P05179/P08683/P11510/P20814/P10633/P10634/P11715
## GO:0020037 P04762/P17178/P20816/P18125/P15149/P00176/P13107/P05178/P05179/P08683/P11510/P20814/P19225/P10633/P10634/P12939/P11715/P01946
## GO:0046906 P04762/P17178/P20816/P18125/P15149/P00176/P13107/P05178/P05179/P08683/P11510/P20814/P19225/P10633/P10634/P12939/P11715/P01946
## GO:0005506 P24483/P17178/P20816/P18125/P15149/P00176/P13107/P05179/P08683/P11510/P20814/P10633/P10634/P11715/P18418/P21816
## GO:0070330 P15149/P00176/P13107/P05179/P08683/P11510/P20814/P10633/P10634
## geneID_symbol
## GO:0004497 Fdx1/Cyp27a1/Cyp3a1/Cyp4a2/Cyp7a1/Cyp2a2/Cyp2b1/Cyp2b3/Cyp2c6/Cyp2c7/Cyp2c11/Cyp2c12/Cyp2c13/Cyp2c70/Cyp2d1/Cyp2d2/Cyp2d10/Cyp17a1/Akr1c9
## GO:0008395 Fdx1/Cyp27a1/Cyp3a1/Cyp7a1/Cyp2a2/Cyp2b1/Cyp2b3/Cyp2c7/Cyp2c11/Cyp2c12/Cyp2c13/Cyp2d1/Cyp2d2/Cyp17a1
## GO:0020037 Cat/Cyp27a1/Cyp4a2/Cyp7a1/Cyp2a2/Cyp2b1/Cyp2b3/Cyp2c6/Cyp2c7/Cyp2c11/Cyp2c12/Cyp2c13/Cyp2c70/Cyp2d1/Cyp2d2/Cyp2d10/Cyp17a1/Hba1
## GO:0046906 Cat/Cyp27a1/Cyp4a2/Cyp7a1/Cyp2a2/Cyp2b1/Cyp2b3/Cyp2c6/Cyp2c7/Cyp2c11/Cyp2c12/Cyp2c13/Cyp2c70/Cyp2d1/Cyp2d2/Cyp2d10/Cyp17a1/Hba1
## GO:0005506 Fdx1/Cyp27a1/Cyp4a2/Cyp7a1/Cyp2a2/Cyp2b1/Cyp2b3/Cyp2c7/Cyp2c11/Cyp2c12/Cyp2c13/Cyp2d1/Cyp2d2/Cyp17a1/Calr/Cdo1
## GO:0070330 Cyp2a2/Cyp2b1/Cyp2b3/Cyp2c7/Cyp2c11/Cyp2c12/Cyp2c13/Cyp2d1/Cyp2d2
## Count FoldEnrich RichFactor
## GO:0004497 19 20.83636 0.11515152
## GO:0008395 14 39.58224 0.21875000
## GO:0020037 18 18.19582 0.10055866
## GO:0046906 18 17.32475 0.09574468
## GO:0005506 16 13.91903 0.07692308
## GO:0070330 9 39.72015 0.219512208.2.4 Some features
We could get conclude some features through examples above:
- No need to sepcify input ID type and organism. The genekitr will automatically detect organism name and convert ID to suitable one.
- Return a data.frame object which is easy to filter or export
- Non-symbol IDs will be automatically converted to the latest gene symbol to easily understand
- If input symbols are mixed with aliases, a new column “geneID_symbol” will be added; if all symbols are official, only “geneID” column will return
-
genORA()includes five gene-level measurement: “Count”, “BgRatio”,“GeneRatio”, “FoldEnrichment” and “RichFactor”.-
BgRatio: Number of all genes in specific term / Number of universal genes (if not specified, is all genes in gene set) -
GeneRatio: Number of genes enriched in specific term / Number of input genes -
FoldEnrichment: GeneRatio / BgRatio -
RichFactor: Number of genes enriched in specific term / Number of all genes in specific term
-
8.3 Advanced usage
8.3.1 Additional arguments
-
pAdjustMethod: choose from “holm”, “hochberg”, “hommel”, “bonferroni”, “BH”, “BY”, “fdr”, “none” -
min_gset_size: Minimal size of each gene set for analysis. Default is 10. Pathway with less than ten genes will be omitted (e.g. GO:0062196 is a child term with 8-gene in the bottom of the directed acyclic graph). -
max_gset_size: Maximal size. Default is 500. The pathway will more than 500 genes will be omitted (e.g. GO:0007049 has 1796 genes as parent nodes at the top of the directed acyclic graph). -
universe: If uses have their background genes, set them asuniverse. If missing, all genes in the geneset will be used as background universe.
8.3.2 Group enrichment
If there is multi-group comparison (e.g. multiple experimental conditions or multiple time points), user could specify gene groups as the group_list argument:
For example, we have 100 genes:
- 50/100 genes are up-regulated and 50/100 are down-regulated
- 40/100 genes are detected at time point 1 and 60/100 are detected at time point 2
id_100 <- c(head(names(geneList), 50), tail(names(geneList), 50))
two_groups <- list(
exp_group = c(rep("up", 50), rep("down", 50)),
time_group = c(rep("time1", 40), rep("time2", 60))
)
gora <- genORA(id_100, geneset = gs, group_list = two_groups)
head(gora)## ID Cluster Description GeneRatio
## 1 DOID:0050774 down.time2 Rapadilino syndrome 0.1020408
## 2 DOID:0050427 down.time2 Xeroderma pigmentosum 0.1836735
## 3 DOID:0060535 down.time2 Warsaw breakage syndrome 0.1020408
## 4 DOID:5683 down.time2 Hereditary breast ovarian cancer syndrome 0.1632653
## 5 DOID:2717 down.time2 Bloom syndrome 0.1224490
## 6 DOID:7400 down.time2 Nijmegen breakage syndrome 0.1020408
## BgRatio pvalue p.adjust qvalue
## 1 24/19143 3.649969e-09 2.522129e-06 1.917194e-06
## 2 281/19143 3.421889e-08 1.182263e-05 8.986961e-06
## 3 54/19143 2.563977e-07 5.905694e-05 4.489209e-05
## 4 274/19143 4.315571e-07 7.455149e-05 5.667026e-05
## 5 137/19143 1.307128e-06 1.806450e-04 1.373172e-04
## 6 107/19143 7.786564e-06 8.967526e-04 6.816659e-04
## geneID
## 1 641/5888/8318/9156/55388
## 2 641/5888/595/7516/8318/993/9156/63967/51514
## 3 113130/157570/5888/8318/63967
## 4 29128/641/5888/595/7516/9156/374/3690
## 5 54821/641/5888/7516/9156/63967
## 6 157570/641/5888/7516/9156
## geneID_symbol Count FoldEnrich RichFactor
## 1 BLM/RAD51/CDC45/EXO1/MCM10 5 81.39031 0.20833333
## 2 BLM/RAD51/CCND1/XRCC2/CDC45/CDC25A/EXO1/CLSPN/DTL 9 12.51267 0.03202847
## 3 CDCA5/ESCO2/RAD51/CDC45/CLSPN 5 36.17347 0.09259259
## 4 UHRF1/BLM/RAD51/CCND1/XRCC2/EXO1/AREG/ITGB3 8 11.40652 0.02919708
## 5 ERCC6L/BLM/RAD51/XRCC2/EXO1/CLSPN 6 17.10979 0.04379562
## 6 ESCO2/BLM/RAD51/XRCC2/EXO1 5 18.25577 0.04672897table(gora$Cluster)##
## down.time2
## 608.3.3 Add background genes
With the help of transId(), background genes could be different type from input IDs.
For example, the input genes are symbols while the background genes are Entrez.
## [1] "948" "1638" "158471" "10610" "6947" "100133941"8.3.4 [GO specific] Simplify GO ORA result
To improve GO term simplifying performance, genekitr extracts species-specific GO term information from Bioconductor organism-level annotation packages for fifteen species, including human, mouse, rat, fly, thale cress, yeast, zebrafish, worm, bovine, pig, chicken, mosquito, dog, frog, and chimpanzee.
Then genekitr obtains the ancestors of GO terms and their relations from GO.db
To calculate semantic similarity for GO BP, CC and MF, genekitr utilizes five algorithms (‘Resnik’, ‘Lin’, ‘Jiang’, ‘Rel’ and ‘Wang’) from GOSemSim
## [1] 34 11## [1] 22 11Let’s look which terms are considered as redundant:
## [1] "GO:0044389" "GO:0140666" "GO:0001091" "GO:0140296" "GO:0005507"
## [6] "GO:0001099"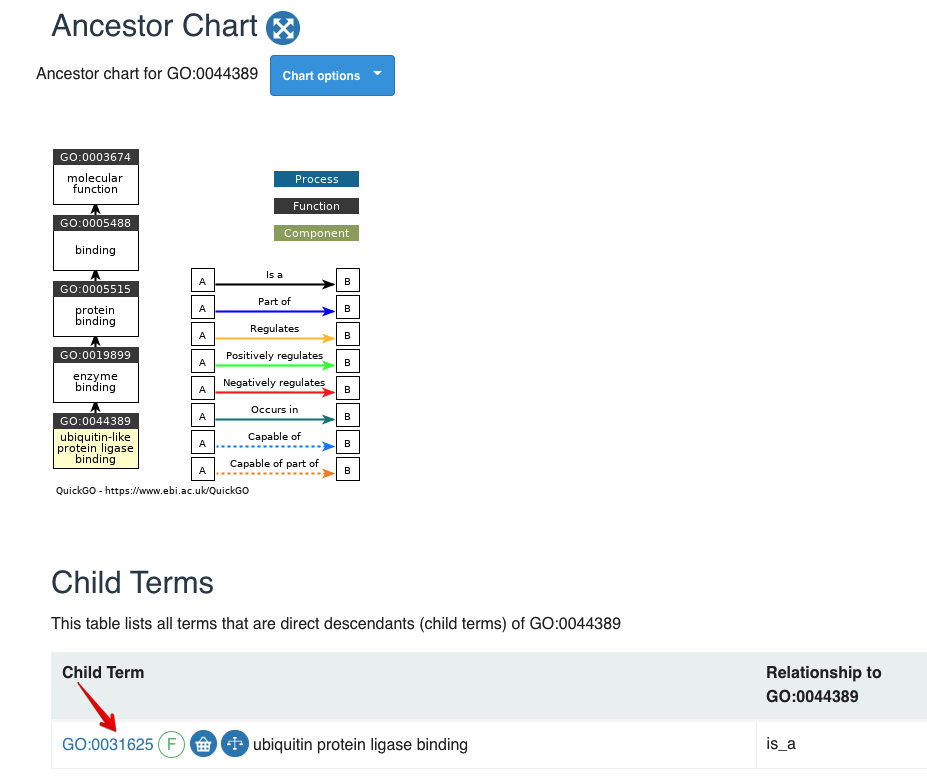
Figure 8.1: Term GO:0044389
The first record GO:0044389 has the child term of GO:0031625, so we only remain the child.
8.4 Comparison with other tools
8.4.1 genekitr::genORA vs clusterProfiler::enrichGO/enrichKEGG/enrichDO...
The function genORA does not require to load the orgdb ahead and no need to specify input ID type.
Next, take GO analysis as example:
The input IDs are mixture of official symbols and aliases. Also all the five genes are very popular in biomedical research.
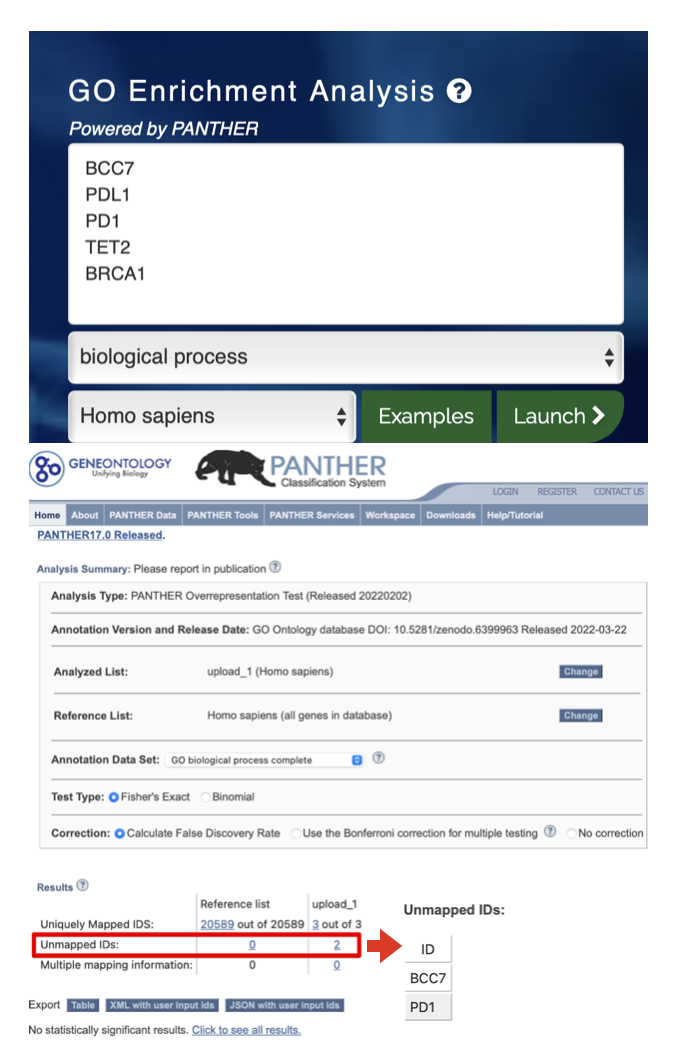
check_gene <- c("BCC7", "PDL1", "PD1", "TET2", "BRCA1")
# Genekitr
start = Sys.time()
gs <- geneset::getGO(org = "human",ont = "bp")
genekitr_res <- genekitr::genORA(check_gene, gs, p_cutoff = 0.01, q_cutoff = 0.01)
end = Sys.time()
(end-start)## Time difference of 7.568743 secs# clusterProfiler
start = Sys.time()
require(org.Hs.eg.db)
clustp_res <- clusterProfiler::enrichGO(check_gene,
OrgDb = "org.Hs.eg.db",
keyType = "SYMBOL",
pvalueCutoff = 0.01,
qvalueCutoff = 0.01,
ont = "BP")
end = Sys.time()
(end-start)## Time difference of 43.37098 secsThen check the result:
class(genekitr_res)## [1] "data.frame"class(clustp_res)## [1] "enrichResult"
## attr(,"package")
## [1] "DOSE"isS4(clustp_res)## [1] TRUE# we need to convert the S4 to dataframe to view
clustp_res2 <- as.data.frame(clustp_res)
dim(genekitr_res)## [1] 24 12dim(clustp_res2)## [1] 11 9head(genekitr_res,5)## Hs_BP_ID Description
## GO:0070234 GO:0070234 positive regulation of T cell apoptotic process
## GO:0070230 GO:0070230 positive regulation of lymphocyte apoptotic process
## GO:2000108 GO:2000108 positive regulation of leukocyte apoptotic process
## GO:0070232 GO:0070232 regulation of T cell apoptotic process
## GO:0070231 GO:0070231 T cell apoptotic process
## GeneRatio BgRatio pvalue p.adjust qvalue
## GO:0070234 0.6 13/18957 2.517291e-09 1.522961e-06 4.107159e-07
## GO:0070230 0.6 17/18957 5.983272e-09 1.809940e-06 4.881090e-07
## GO:2000108 0.6 27/18957 2.571649e-08 5.186160e-06 1.398616e-06
## GO:0070232 0.6 34/18957 5.258194e-08 7.953019e-06 2.144790e-06
## GO:0070231 0.6 50/18957 1.720087e-07 2.081305e-05 5.612915e-06
## geneID geneID_symbol Count FoldEnrich RichFactor
## GO:0070234 BCC7/PDL1/PD1 TP53/CD274/PDCD1 3 874.9385 0.23076923
## GO:0070230 BCC7/PDL1/PD1 TP53/CD274/PDCD1 3 669.0706 0.17647059
## GO:2000108 BCC7/PDL1/PD1 TP53/CD274/PDCD1 3 421.2667 0.11111111
## GO:0070232 BCC7/PDL1/PD1 TP53/CD274/PDCD1 3 334.5353 0.08823529
## GO:0070231 BCC7/PDL1/PD1 TP53/CD274/PDCD1 3 227.4840 0.06000000head(clustp_res,5)## ID Description GeneRatio BgRatio
## GO:0051568 GO:0051568 histone H3-K4 methylation 2/2 57/18866
## GO:0044728 GO:0044728 DNA methylation or demethylation 2/2 98/18866
## GO:0034968 GO:0034968 histone lysine methylation 2/2 116/18866
## GO:0006304 GO:0006304 DNA modification 2/2 120/18866
## GO:0018022 GO:0018022 peptidyl-lysine methylation 2/2 132/18866
## pvalue p.adjust qvalue geneID Count
## GO:0051568 8.968633e-06 0.001892382 9.440666e-06 TET2/BRCA1 2
## GO:0044728 2.670922e-05 0.002006407 1.000951e-05 TET2/BRCA1 2
## GO:0034968 3.748169e-05 0.002006407 1.000951e-05 TET2/BRCA1 2
## GO:0006304 4.012283e-05 0.002006407 1.000951e-05 TET2/BRCA1 2
## GO:0018022 4.858571e-05 0.002006407 1.000951e-05 TET2/BRCA1 2-
clustp_resonly contains two genes:TET2andBRCA1 -
genekitr_resincludes all five genes and gives mapping for aliases
If you look into deeper, you may find eight terms are unique in clustp_res, such as GO:0016571:
clustp_res$ID[!clustp_res$ID%in%genekitr_res$Hs_BP_ID ]## [1] "GO:0034968" "GO:0018022" "GO:0016571" "GO:0006479" "GO:0008213"
## [6] "GO:0043414" "GO:0032259" "GO:0018205"Question1: Why some terms are omited in genGO result?
To get adjusted p value, the critical parameter is the number of statistical testing.
For simple explanation, adj.p = f * p (f>1) and f is related with testing methods and the number of tests. genORA recognizes five genes so we need to do more testings (for example, we do five testings) while enrichGO only includes two genes so the testing number will be less. This directly reduces the adj.p in enrichGO result and it could be remained under the threshold 0.01.
So, if we set cutoff higher(e.g. 0.05), genekitr could also get those terms:
genekitr_res2 <- genekitr::genORA(check_gene, gs, p_cutoff = 0.05, q_cutoff = 0.05)
table(clustp_res$ID%in%genekitr_res2$Hs_BP_ID)##
## TRUE
## 11Question2: Why enrichGO only recognize two genes?
We all know that it is not possible for such essential genes (“BCC7”, “PD1” or “PDL1”) without enriched terms.
The core reason is that the three genes are actually aliases while enrichGO sets argument keyType = "SYMBOL", which cause enrichGO only accepts official symbols. This function is based on organism annotation package (e.g. org.Hs.eg.db) and it will accept different types of data from org.Hs.eg.db:
## [1] "ACCNUM" "ALIAS" "ENSEMBL" "ENSEMBLPROT" "ENSEMBLTRANS"
## [6] "ENTREZID" "ENZYME" "EVIDENCE" "EVIDENCEALL" "GENENAME"
## [11] "GO" "GOALL" "IPI" "MAP" "OMIM"
## [16] "ONTOLOGY" "ONTOLOGYALL" "PATH" "PFAM" "PMID"
## [21] "PROSITE" "REFSEQ" "SYMBOL" "UCSCKG" "UNIGENE"
## [26] "UNIPROT"Only by specifying keyType = 'ALIAS', the program could recognize gene alias, however it still could not distinguish the mixture of symbol and alias.