11 Plot ORA
After selecting interested terms or pathways from genORA or genGSEA result, user could pass the data frame to plotEnrich, which includes many ready-made plot types, including barplot, dotplot, heatmap, wego-like plot, chord plot, network, wordcloud etc.
It is worth mentioning that almost all plots are based on ggplot2 and plot_theme function could easily change their border, legend, label etc., which helps user make own plot.
11.1 Get ORA result
For more details, please refer to chapter8
# 1st step: prepare input IDs
data(geneList, package = "genekitr")
entrez_id <- names(geneList)[abs(geneList) > 2]
head(entrez_id, 5)## [1] "948" "1638" "158471" "10610" "6947"# 2nd step: prepare gene set
hg_gs <- geneset::getGO(org = "human",ont = "bp")
# 3rd step: ORA analysis
ego <- genORA(entrez_id, geneset = hg_gs, p_cutoff = 0.01, q_cutoff = 0.01)
# next we only show ten sample terms
ego <- ego[1:10, ]
head(ego)## Hs_BP_ID
## GO:0050673 GO:0050673
## GO:0071900 GO:0071900
## GO:0050678 GO:0050678
## GO:0010165 GO:0010165
## GO:0043405 GO:0043405
## GO:1901992 GO:1901992
## Description GeneRatio
## GO:0050673 epithelial cell proliferation 0.128
## GO:0071900 regulation of protein serine/threonine kinase activity 0.112
## GO:0050678 regulation of epithelial cell proliferation 0.104
## GO:0010165 response to X-ray 0.040
## GO:0043405 regulation of MAP kinase activity 0.072
## GO:1901992 positive regulation of mitotic cell cycle phase transition 0.056
## BgRatio pvalue p.adjust qvalue
## GO:0050673 443/18957 3.656558e-08 0.0001006285 7.736508e-05
## GO:0071900 372/18957 1.680273e-07 0.0002312055 1.777552e-04
## GO:0050678 382/18957 1.471777e-06 0.0013501101 1.037990e-03
## GO:0010165 32/18957 2.008272e-06 0.0013816909 1.062270e-03
## GO:0043405 183/18957 3.495297e-06 0.0019238117 1.479063e-03
## GO:1901992 100/18957 4.408629e-06 0.0020220911 1.554622e-03
## geneID
## GO:0050673 5176/26471/3488/6591/8549/3725/6657/4208/407007/6422/29128/595/7039/374/3690/10468
## GO:0071900 100133941/8654/5027/10125/6422/641/595/7039/7099/993/9134/57214/81848/10253
## GO:0050678 5176/26471/6591/3725/6657/4208/407007/6422/29128/595/7039/374/3690
## GO:0010165 6422/641/5888/595/7516
## GO:0043405 100133941/8654/5027/10125/6422/7039/7099/81848/10253
## GO:1901992 407007/113130/6241/595/407006/993/51514
## geneID_symbol
## GO:0050673 SERPINF1/NUPR1/IGFBP5/SNAI2/LGR5/JUN/SOX2/MEF2C/MIR222/SFRP1/UHRF1/CCND1/TGFA/AREG/ITGB3/FST
## GO:0071900 CD24/PDE5A/P2RX7/RASGRP1/SFRP1/BLM/CCND1/TGFA/TLR4/CDC25A/CCNE2/CEMIP/SPRY4/SPRY2
## GO:0050678 SERPINF1/NUPR1/SNAI2/JUN/SOX2/MEF2C/MIR222/SFRP1/UHRF1/CCND1/TGFA/AREG/ITGB3
## GO:0010165 SFRP1/BLM/RAD51/CCND1/XRCC2
## GO:0043405 CD24/PDE5A/P2RX7/RASGRP1/SFRP1/TGFA/TLR4/SPRY4/SPRY2
## GO:1901992 MIR222/CDCA5/RRM2/CCND1/MIR221/CDC25A/DTL
## Count FoldEnrich RichFactor
## GO:0050673 16 5.477418 0.03611738
## GO:0071900 14 5.707484 0.03763441
## GO:0050678 13 5.161068 0.03403141
## GO:0010165 5 23.696250 0.15625000
## GO:0043405 9 7.458492 0.04918033
## GO:1901992 7 10.615920 0.0700000011.2 Bar Plot
Bar plot is the simplest way to show enriched terms. The x-axis is enrichment metric (e.g. Gene ratio; Fold enrichment); the y-axis is selected terms. The bar color represents statistical value.
What’s the difference between pvalue and p.adjust?
p.adjusthas seven adjustment methods: https://www.rdocumentation.org/packages/stats/versions/3.5.0/topics/p.adjust. The adjusted p-value is always the p-value, multiplied with some factor: adj.p = f * p The actual size of this factor f depends on the strategy used to correct for multiple testing. By the way, the “q-value” stands for “false discovery rate (FDR)” method.
The basic arguments are:
-
term_metric: The x-axis could be “Count”, “GeneRatio”, “FoldEnrich” or “RichFactor” -
stats_metric: Statistic value of “p.adjust”, “pvalue” or “qvalue” -
up_color: Color for stronger statistic value (e.g. pvalue 0.01) -
down_color: Color for weaker statistic value (e.g. pvalue 1) -
wrap_length: Wrap term text if longer than this number
library(patchwork)
p1 <- plotEnrich(ego, plot_type = "bar")
p2 <- plotEnrich(ego, plot_type = "bar", term_metric = "GeneRatio", stats_metric = "pvalue")
p3 <- plotEnrich(ego, plot_type = "bar", up_color = "#E69056", down_color = "#325CAC")
p4 <- plotEnrich(ego, plot_type = "bar", wrap_length = 25)
p1 + p2 + p3 + p4 + plot_annotation(tag_levels = "A")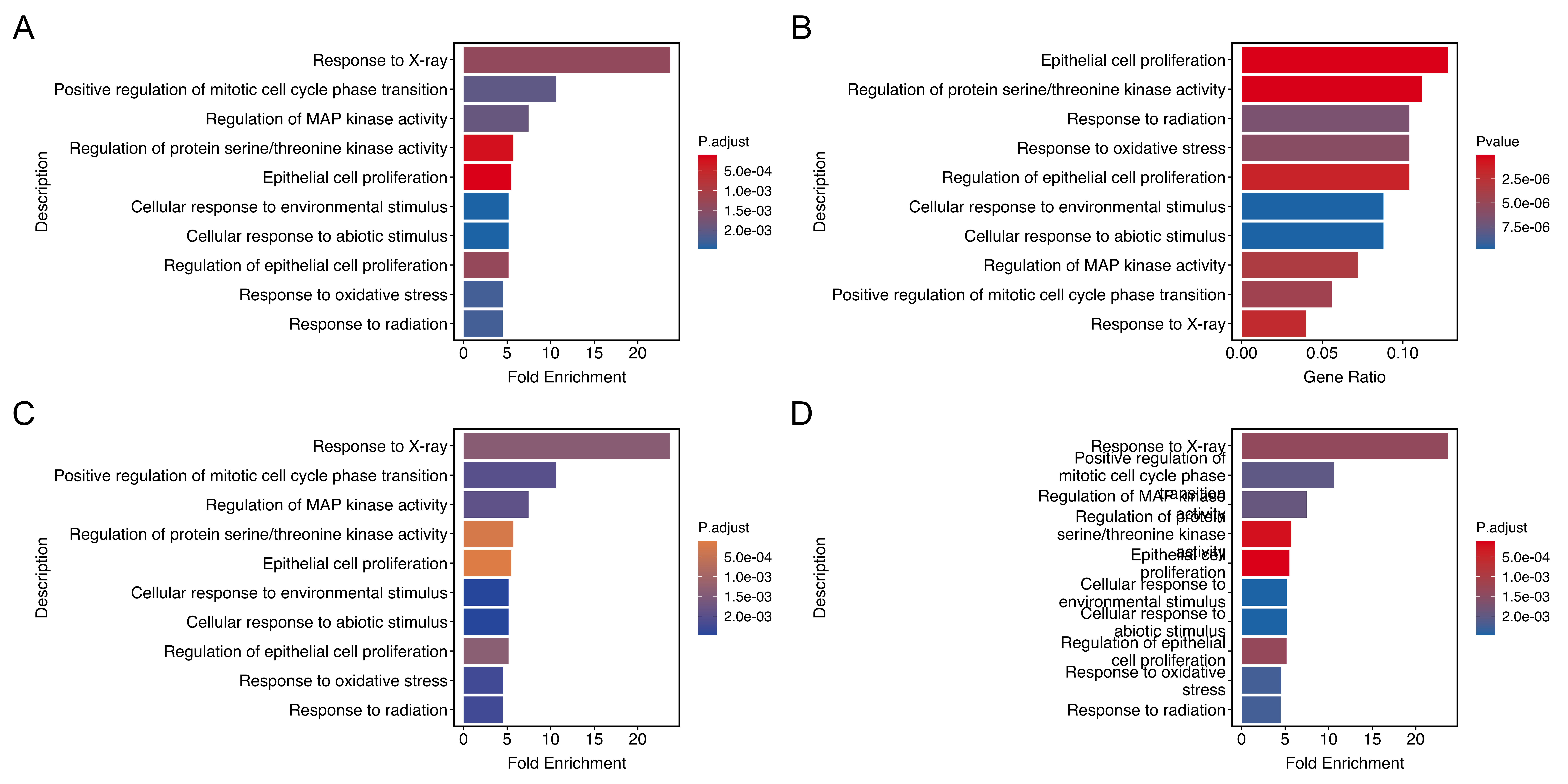
Figure 11.1: Bar plot of enrichment analysis. default (A), modify metrics (B), modify color (C) and modify term length (D).
11.3 Bubble Plot
Enriched gene sets are shown as bubbles with different sizes.
The x-axis is statistic value and the y-axis is “Fold Enrichment”.
The basic arguments are:
-
stats_metric: Statistic value of “p.adjust”, “pvalue” or “qvalue” -
scale_ratio: Change bubble size. Default is 1.
library(patchwork)
p1 <- plotEnrich(ego, plot_type = "bubble")
p2 <- plotEnrich(ego, plot_type = "bubble",
scale_ratio = 0.5, stats_metric = "qvalue")
p1 / p2 + plot_annotation(tag_levels = "A")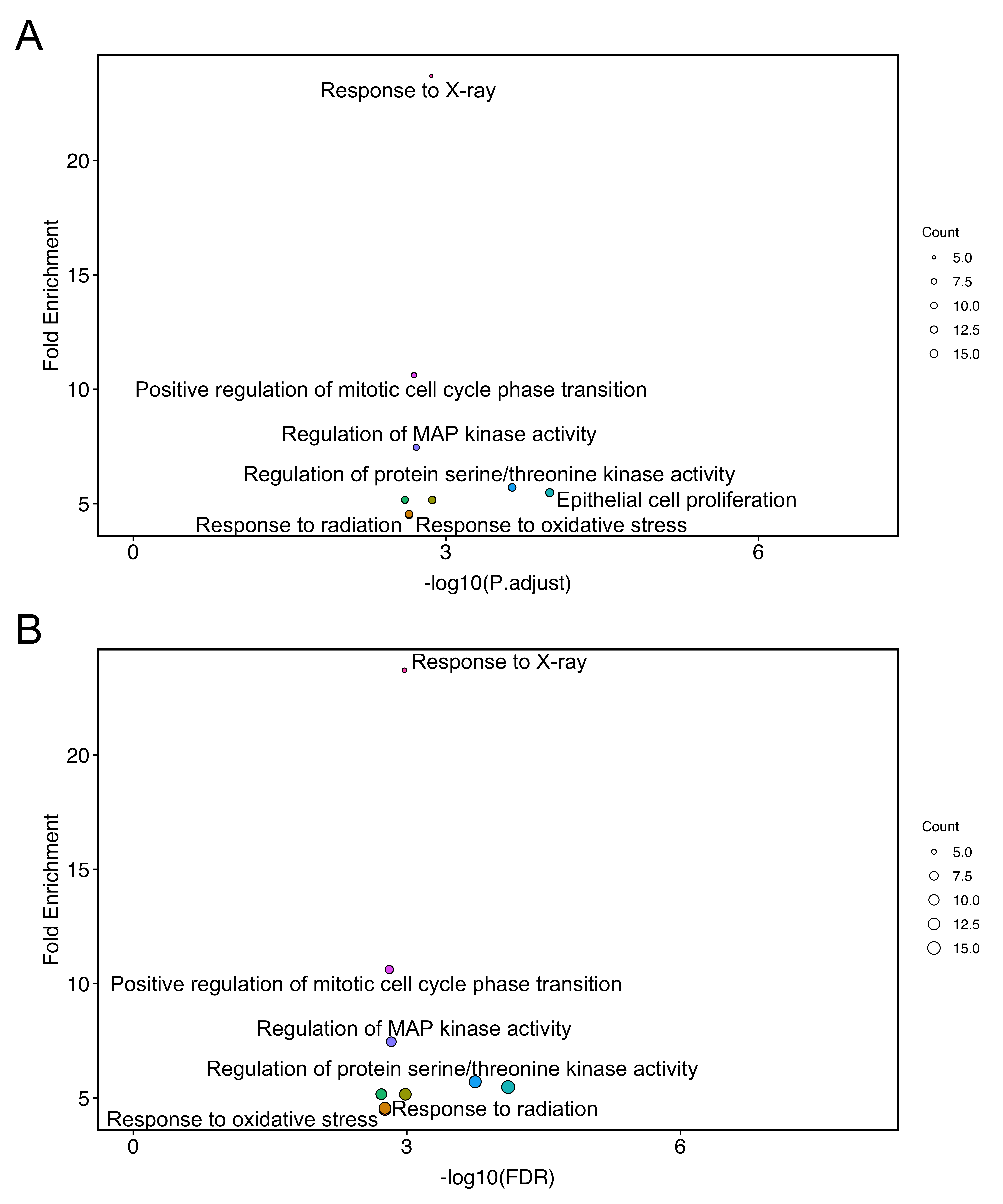
Figure 11.2: Bubble plot of enrichment analysis. default (A), modify bubble size (B).
11.4 Dot Plot
Similar with bar plot, dot plot is also widely used in enrichment analysis plotting. Like bubble plot, dot size represents gene number of enriched term.
library(patchwork)
p1 <- plotEnrich(ego, plot_type = "dot")
p2 <- plotEnrich(ego,
plot_type = "dot",
scale_ratio = 1.5,
stats_metric = "pvalue",
term_metric = "RichFactor"
)
p1 + p2 + plot_annotation(tag_levels = "A")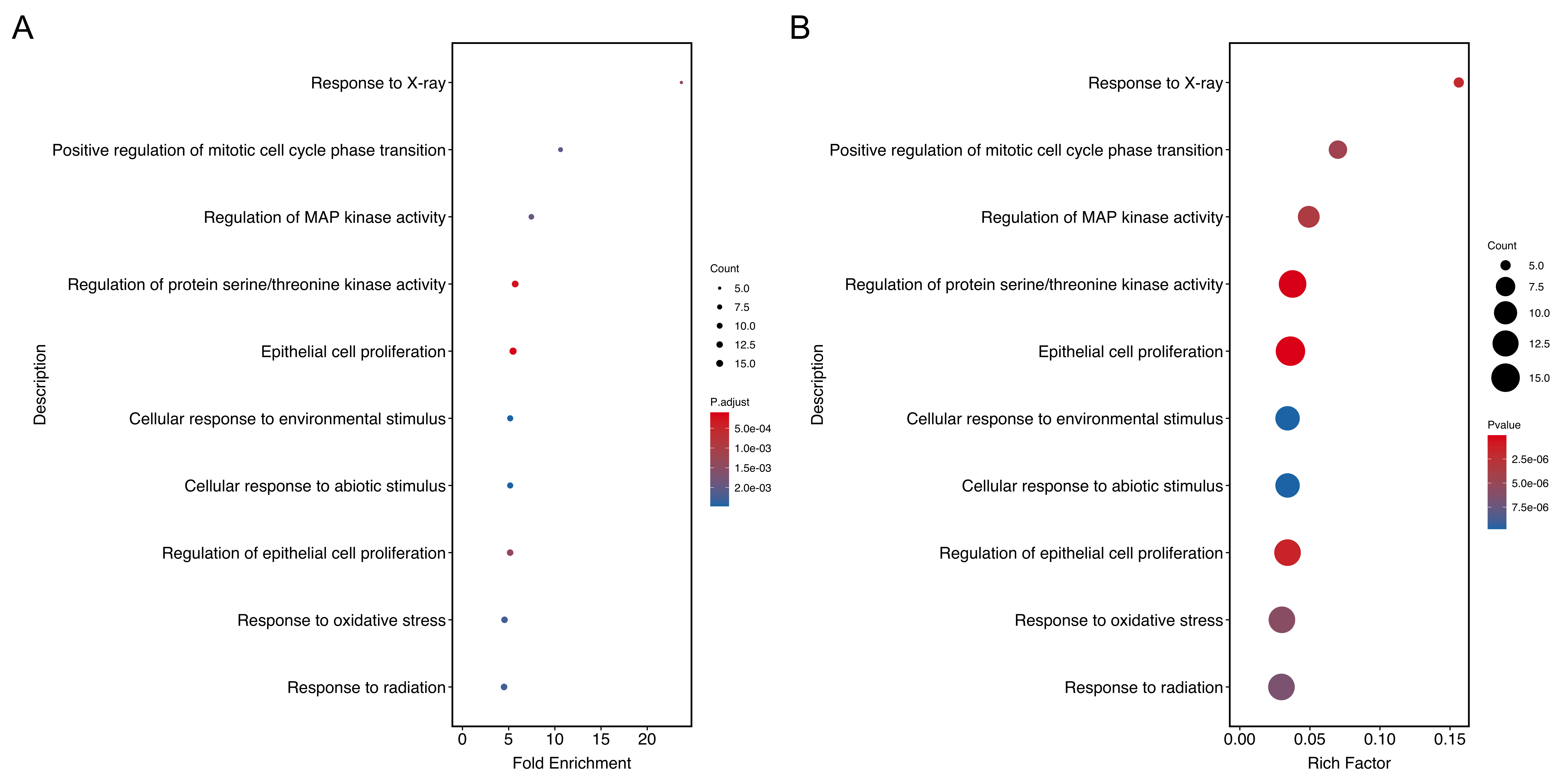
Figure 11.3: Dot plot of enrichment analysis. default (A), modify bubble size (B).
After analyzing group enrichment analysis for ORA, we could use dotplot to show the result:
plotEnrich(gego2,
plot_type = 'dot',
scale_ratio = 2, # dot size
main_text_size = 10,
legend_text_size =8,
n_term = 6) # show terms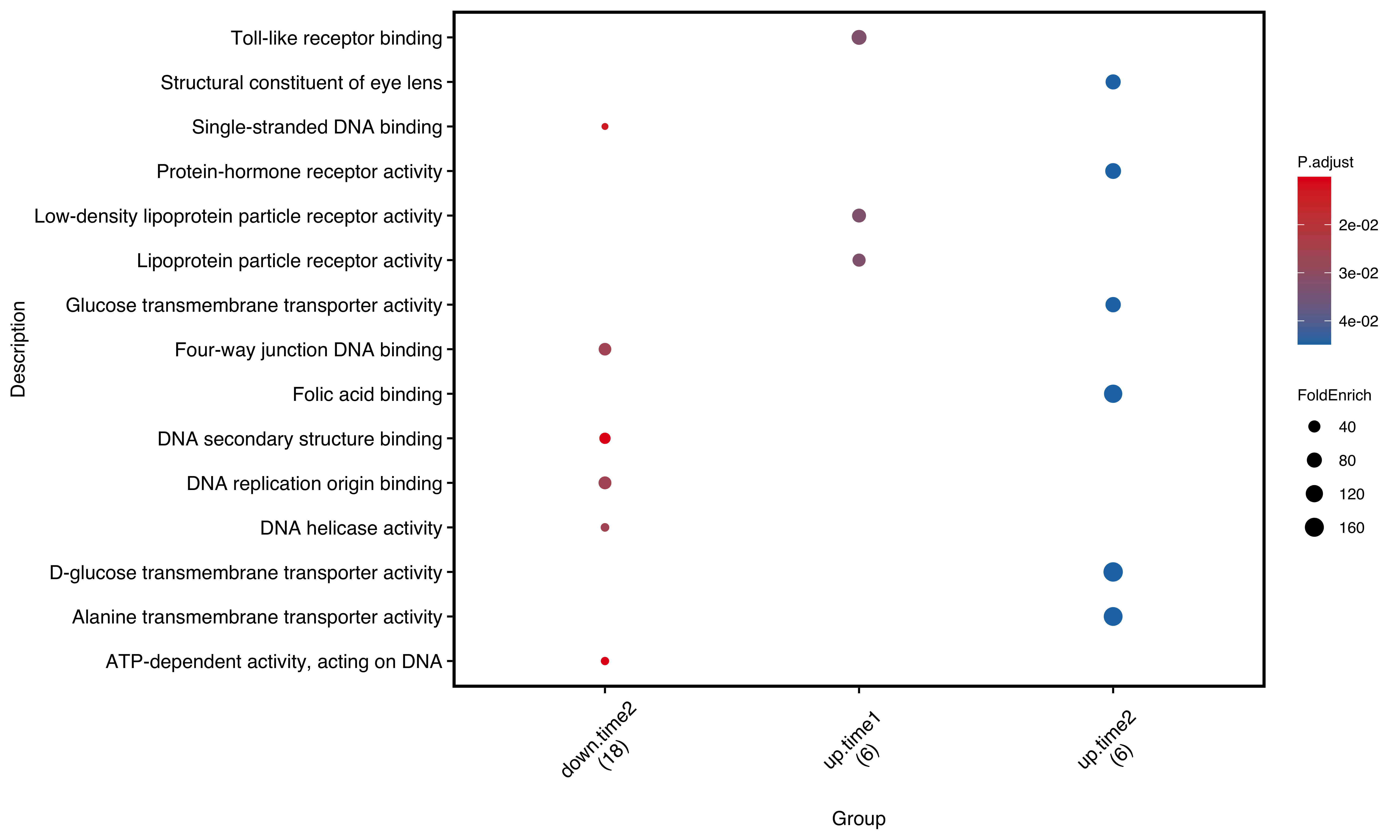
Figure 11.4: Dot plot of group enrichment analysis. number in round brackets shows total gene number in selected pathways
11.5 Lollipop Plot
Lollipop is like combination of barplot and dotplot
library(patchwork)
p1 <- plotEnrich(ego, plot_type = "lollipop")
p2 <- plotEnrich(ego,
plot_type = "lollipop",
scale_ratio = 1.2,
stats_metric = "pvalue",
term_metric = "RichFactor",
up_color = "#a32a31",
down_color = "#f7dcca"
)
p1 + p2 + plot_annotation(tag_levels = "A")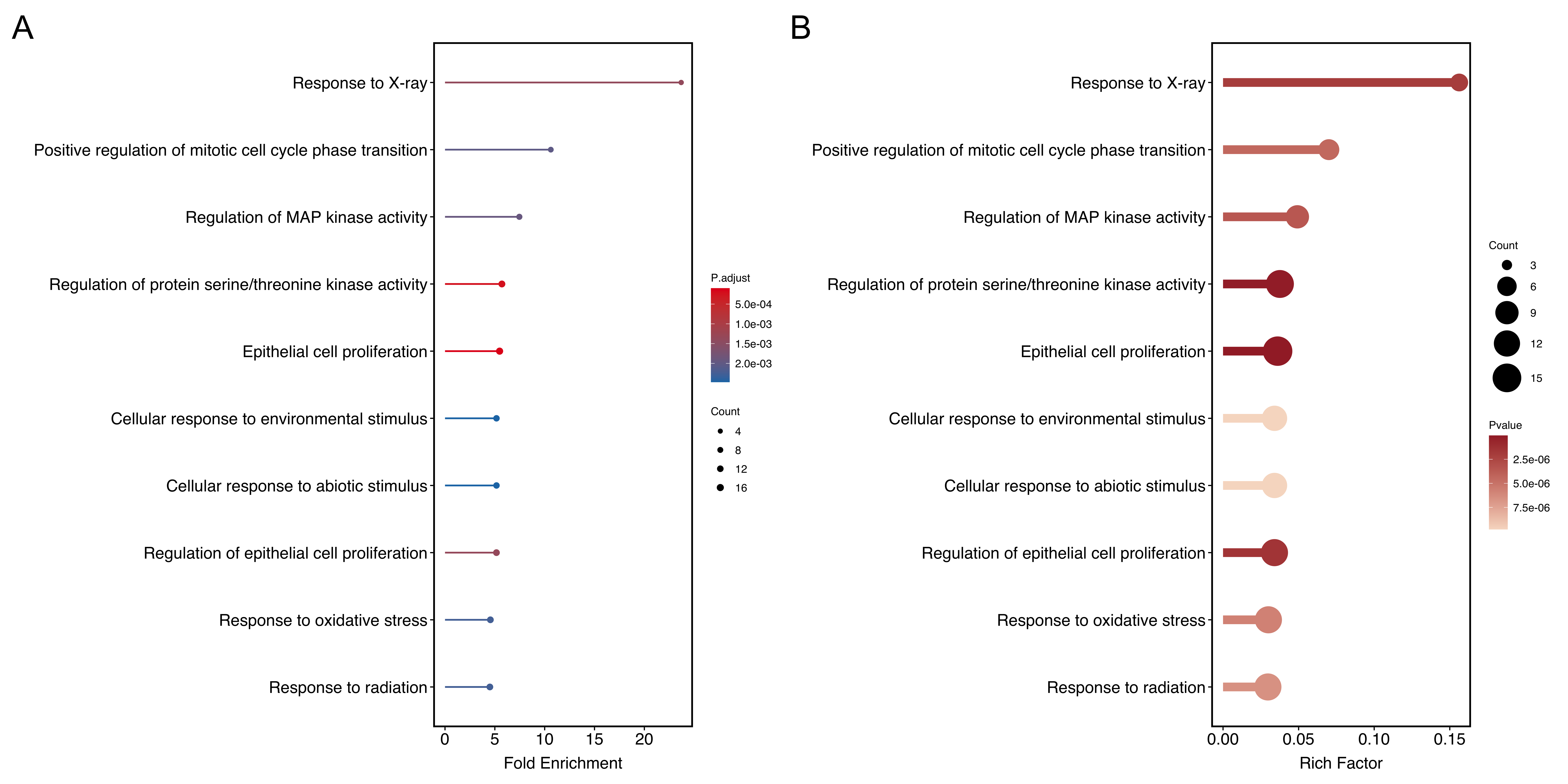
Figure 11.5: Lollipop plot of enrichment analysis. defult (A), selected genes (B)
11.6 Heatmap Plot
Heatmap plot shows interactions between enriched terms and their genes. If fold change is given, heatmap will add color for up and down-regulated genes.
library(patchwork)
p1 <- plotEnrich(ego, plot_type = "geneheat")
show_gene = c('JUN','SOX2','CD24','TLR4')
p2 <- plotEnrich(ego, plot_type = "geneheat", show_gene = show_gene)
p3 <- plotEnrich(ego, plot_type = "geneheat", show_gene = show_gene, fold_change = logfc)
p1 / p2 / p3 + plot_annotation(tag_levels = "A")
Figure 11.6: Heatmap plot of enrichment analysis. all genes (A, default), selected genes (B), selected genes with logFC value (C).
11.7 Chord Plot
Inspired by GOplot, chord plot is reproduced using ggplot2 and it shows similar content with heatmap plot.
-
gene_space: The space between the gene labels and the chord.
library(patchwork)
show_gene = c('JUN','SOX2','CD24','TLR4')
p1 <- plotEnrich(ego, plot_type = "genechord",
show_gene = show_gene) +
ggplot2::theme(legend.position = "none")
p2 <- plotEnrich(ego, plot_type = "genechord",
show_gene = show_gene,
fold_change = logfc,
gene_space = 0.5)
p1 + p2 + plot_annotation(tag_levels = "A")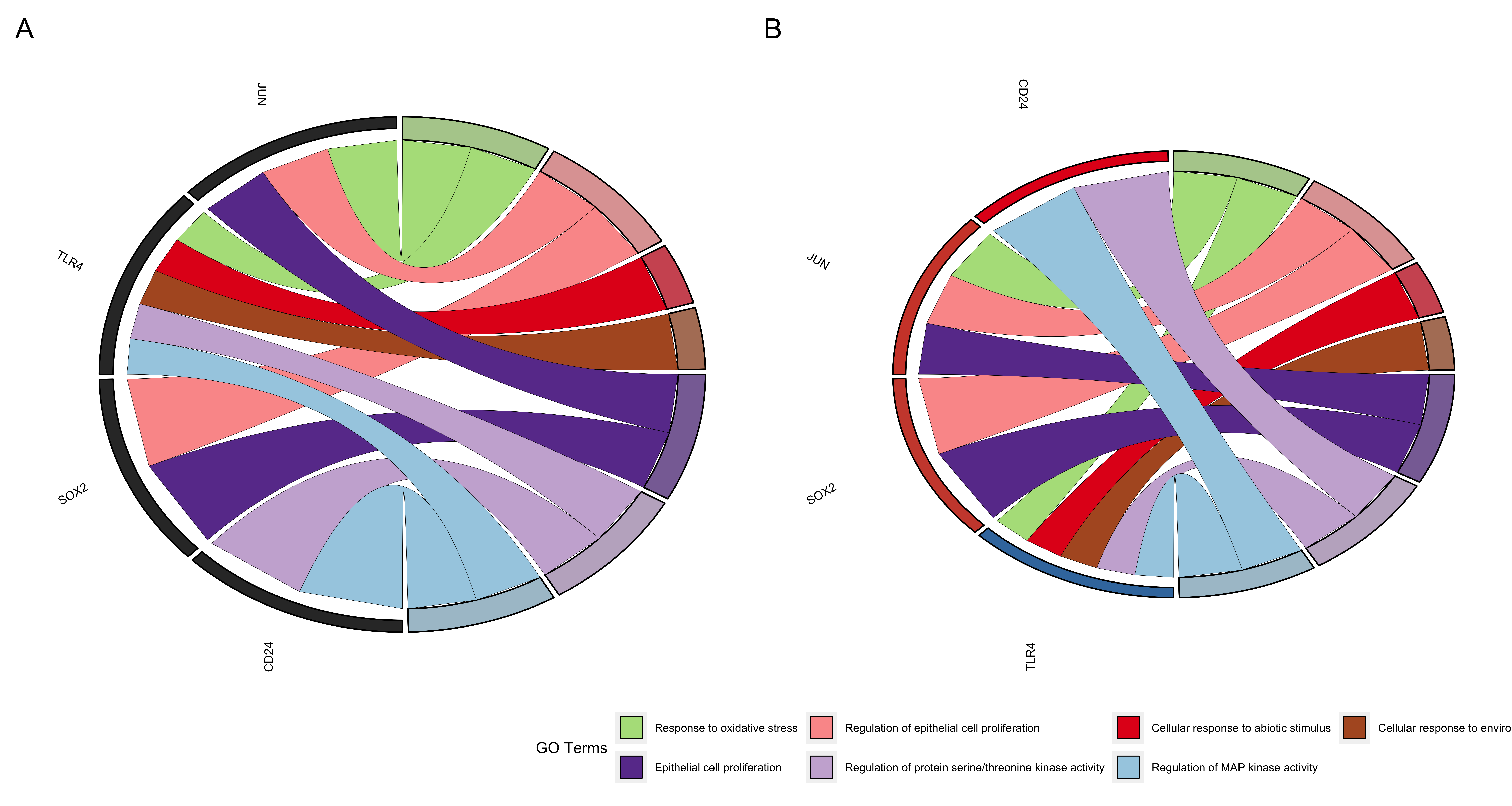
Figure 11.7: Heatmap plot of enrichment analysis. selected genes (A), selected genes with logFC value (B).
11.8 Wordcloud Plot
Wordcloud plot shows term emphasis based on text frequency.
plotEnrich(ego, plot_type = "wordcloud")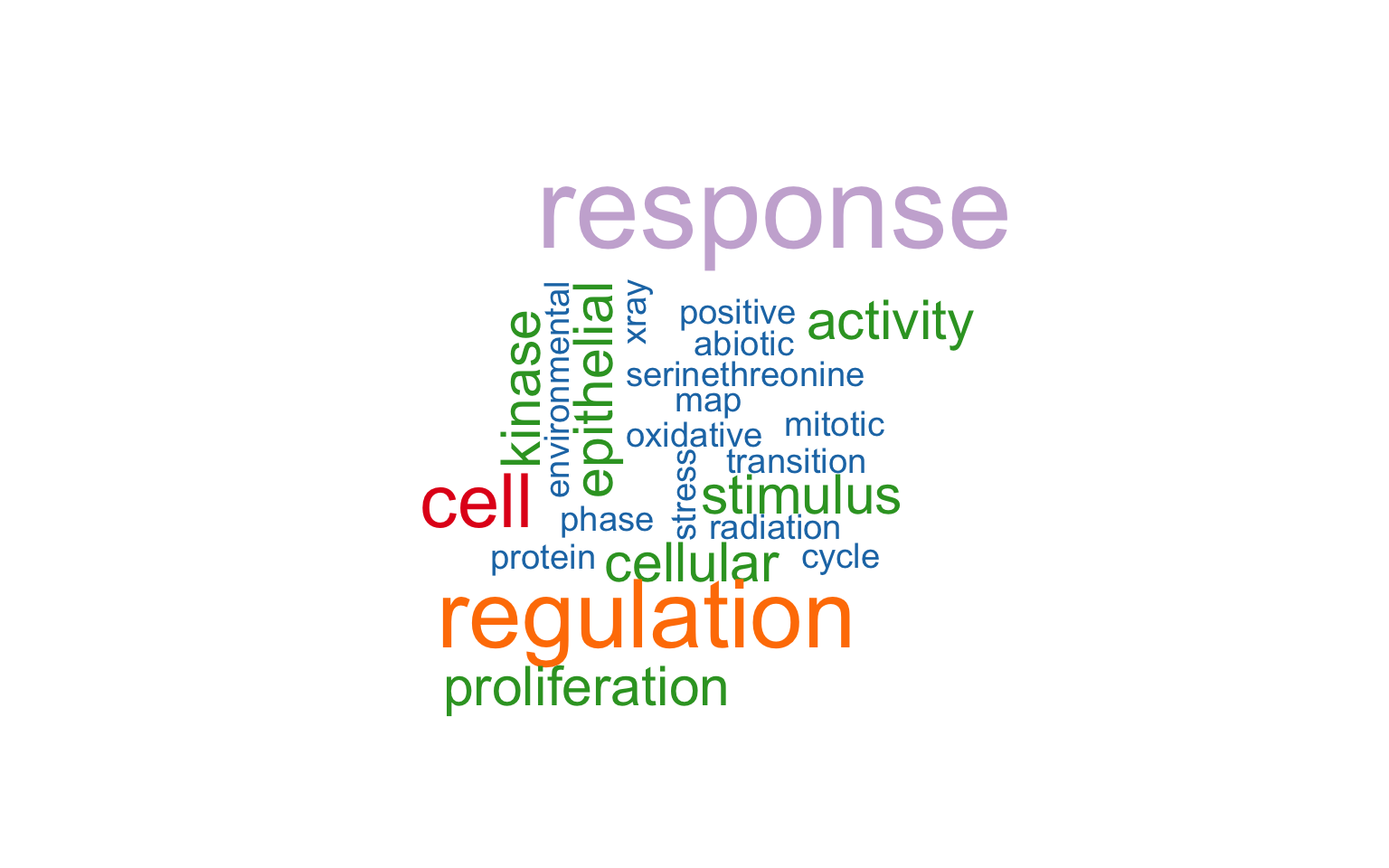
Figure 11.8: Wordcloud plot of enrichment analysis.
11.9 Upset Plot
Inspired by ComplexUpset, the upset plot shows the association between genes and enriched terms. Unlike common Venn diagram, it could emphasize complex relationship among many gene sets.
plotEnrich(ego, plot_type = "upset",main_text_size = 15,legend_text_size = 8)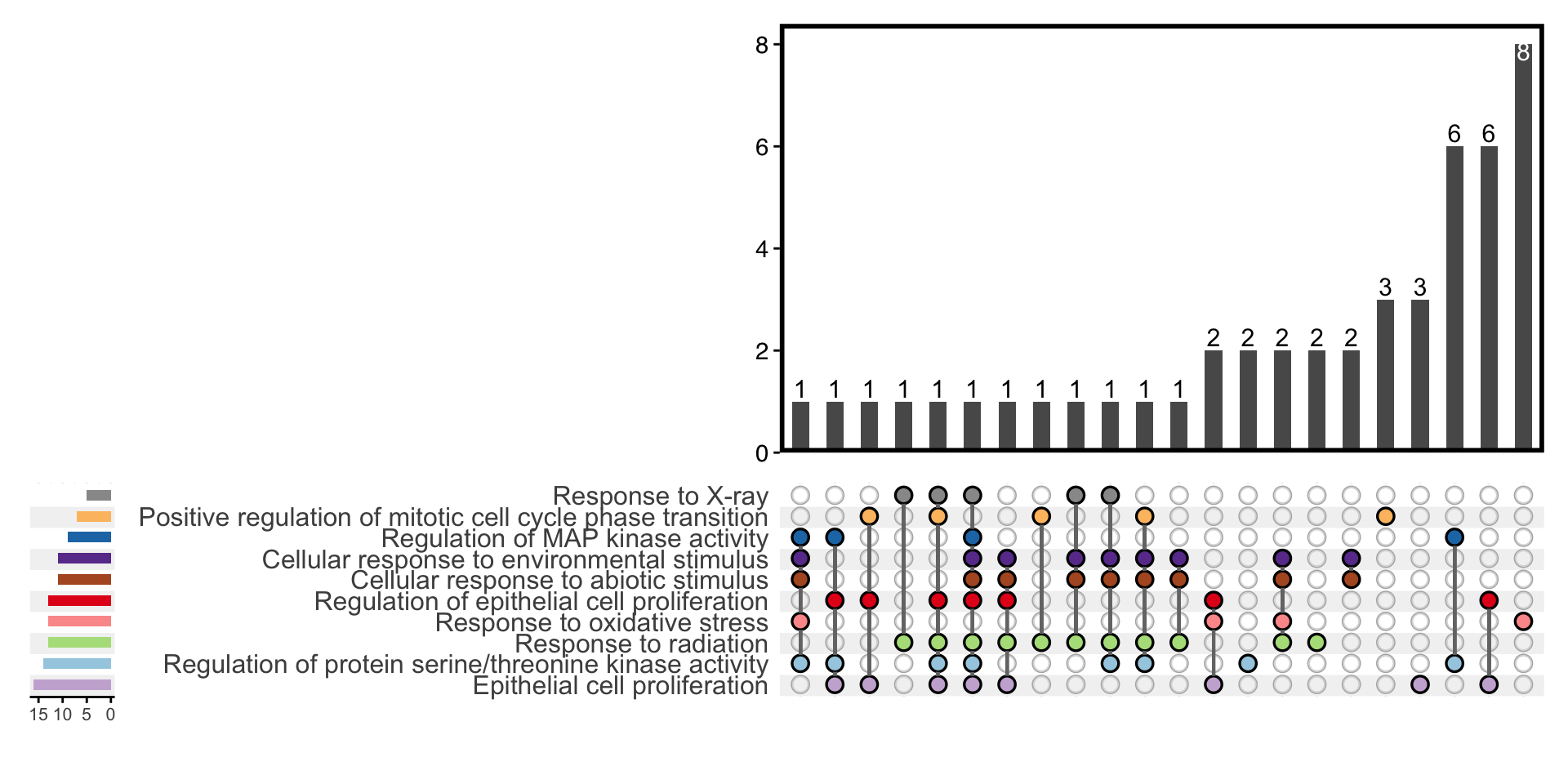
Figure 11.9: Upset plot of enrichment analysis.
11.10 Network Plot
Inspired by enrichplot::emapplot, genekitr reproduced this plot to enhance modification.
Enriched terms are the nodes in the network, the overlapping gene sets are edges. Terms with more overlapped genes are tend to cluster together and the edge will be thicker.
- GO analysis result could use five methods to calculate the similarity between nodes: “Resnik”, “Lin”, “Rel”, “Jiang”, “Wang” and “JC”(Jaccard’s similarity index) methods.
- KEGG only supports “JC” method.
User could define layout argument derived from ggraph, including “nicely” (default), “circle”, “dh”, “drl”, “fr”, “graphopt”, “grid”,“lgl”, “kk”, “mds”, “randomly”, “star” etc.
For more information about the
layout, you could refer to: “Introduction to ggraph: Layouts”
library(patchwork)
library(igraph)
library(ggraph)
p1 <- plotEnrich(ego, plot_type = "network", scale_ratio = 0.5)
p2 <- plotEnrich(ego, plot_type = "network",
layout = "circle", scale_ratio = 0.5)
p3 <- plotEnrich(ego, plot_type = "network",
layout = "grid", sim_method = "Wang",
up_color = "#a32a31", down_color = "#f7dcca")
(p1 + p2) / p3 + plot_annotation(tag_levels = "A")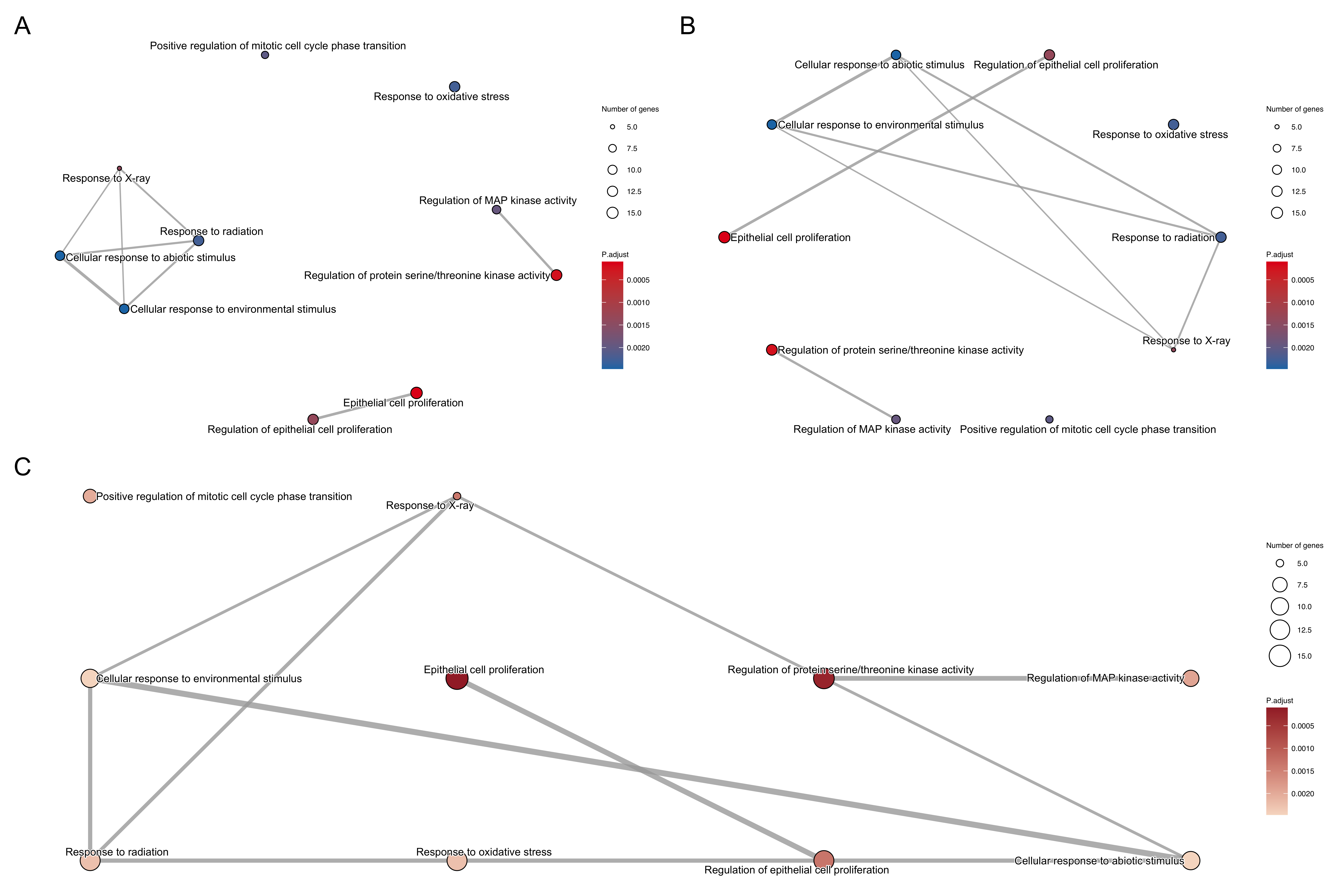
Figure 11.10: Network plot of enrichment analysis. JC method and nicely layout(A, default), circle layout (B), grid layout and Want method (C).
11.11 GO-specific: WEGO Plot
To visulize more than one ontology of GO in one plot, user could utilize
wego plot
Inspired by WEGO, genekitr utilized ggplot2 to reproduce this plot.
Here we generate two ontologies (MF and CC) result.
# 1st step: prepare input IDs
data(geneList, package = "genekitr")
id <- names(geneList)[abs(geneList) > 2]
# 2nd step: prepare CC and MF gene sets
go_cc <- geneset::getGO(org = "human",ont = "cc")
go_mf <- geneset::getGO(org = "human",ont = "mf")
# 3rd step: analysis
ego_cc <- genORA(id, geneset = go_cc)
ego_mf <- genORA(id, geneset = go_mf)
# 4th step: merge two data frames
# Note: each data frame should add new column "Ontology"
ego_cc <- ego_cc %>% dplyr::mutate(Ontology = "cc") %>% dplyr::rename(ID = 1)
ego_mf <- ego_mf %>% dplyr::mutate(Ontology = "mf") %>% dplyr::rename(ID = 1)
all_ego <- rbind(ego_cc,ego_mf)NOTICE: plotEnrich has a parameter n_term for WEGO plot, which specify number of terms. If you want to plot all terms, just set n_term higher.
plotEnrich(all_ego, plot_type = "wego", n_term = 5)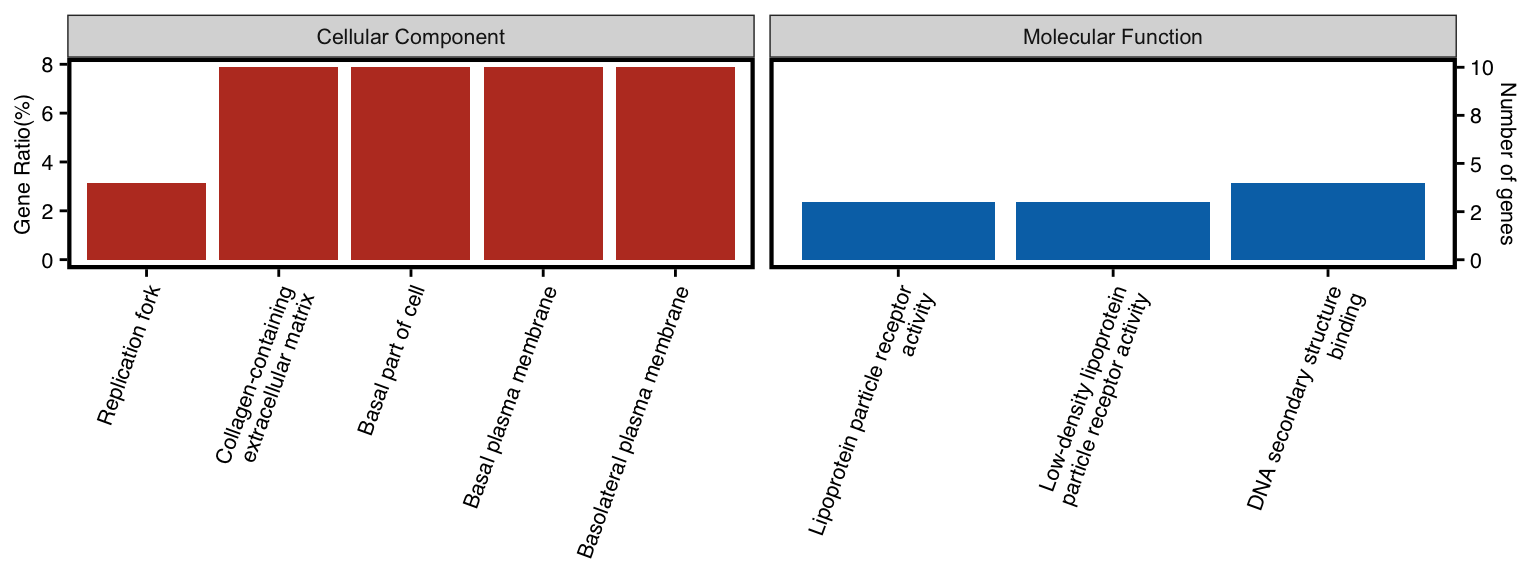
Figure 11.11: WEGO plot of enrichment analysis.
11.12 GO-specific: Map Plot
GO terms are built in a directed acyclic graph with a parent-child relationship. Here the map plot utilized GOSemSim to extract parent and child terms, also utilized ggraph and igraph to draw with default layout “sugiyama”.
To avoid too much unrelated terms messed up, genekitr only shows the closest parent and child with selected terms. Besides, the top three parent term with more edges will be plotted.
library(igraph)
library(ggraph)
plotEnrich(ego, plot_type = "gomap", wrap_length = 25,
up_color = '#a32a31',down_color = '#3665a6')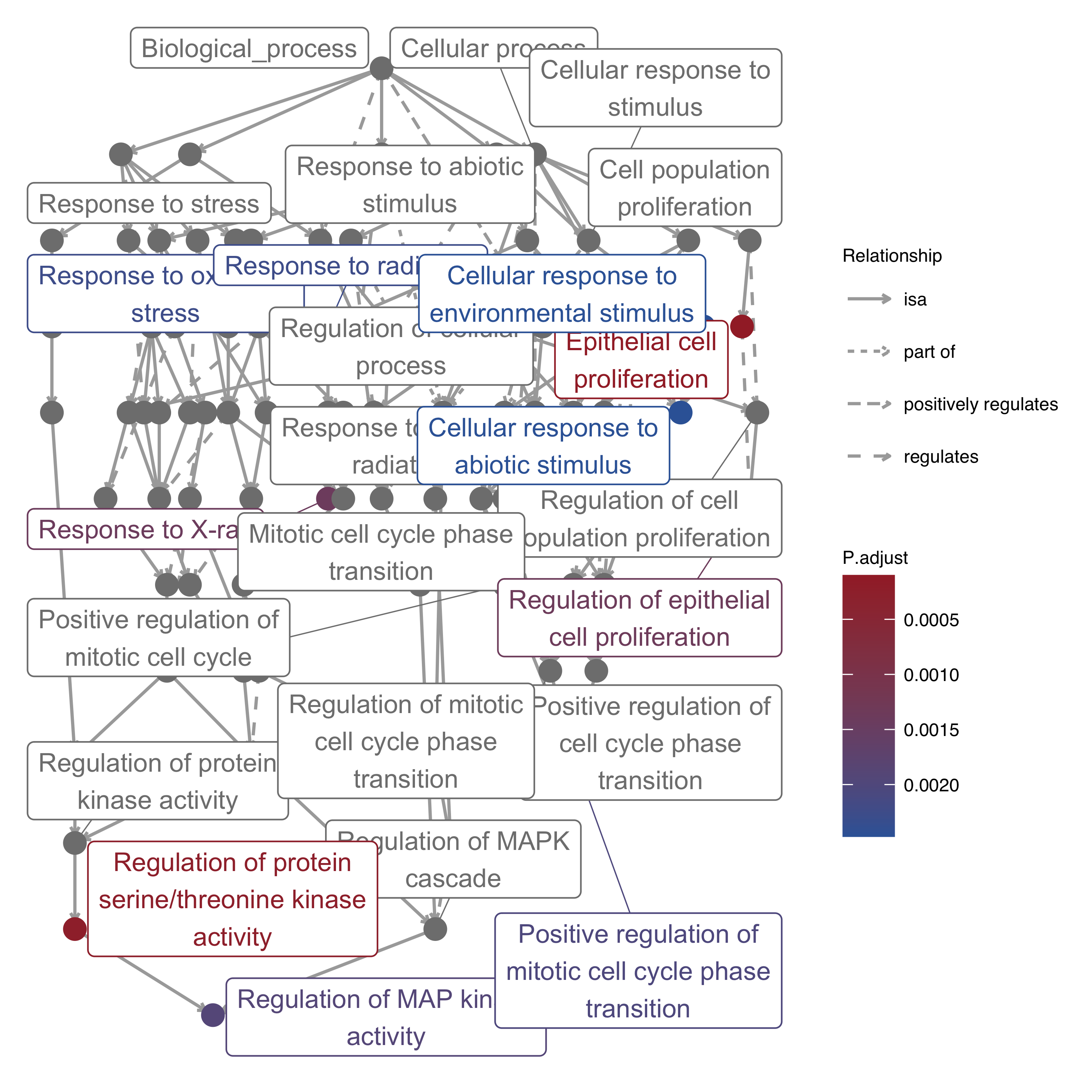
Figure 11.12: Map plot of enrichment analysis.
11.13 GO-specific: Terms Heatmap Plot
Inspired by rrvgo, genekitr scratches main codes to cluster GO terms. Also it could use five methods to calculate the similarity between terms.
plotEnrich(ego, plot_type = "goheat", sim_method = "Rel")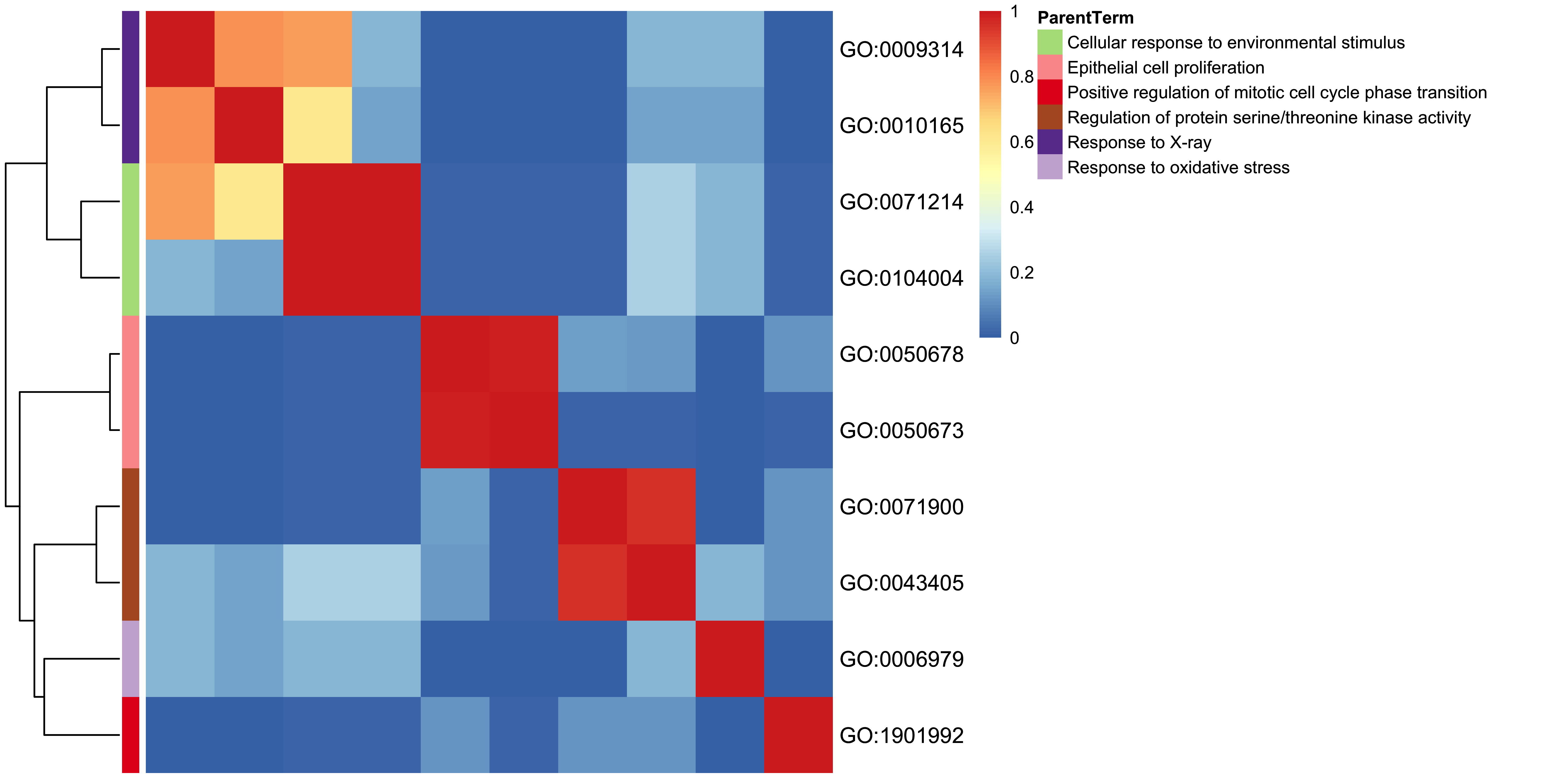
Figure 11.13: GO heatmap plot of enrichment analysis.
11.14 GO-specific: Terms Tangram Plot
According to rrvgo vignettes, tangram plot is space-filling visualization of hierarchical structures. The terms are grouped and colored based on their parent, and the space used by the term is proportional to the score.
plotEnrich(ego, plot_type = "gotangram", sim_method = "Rel")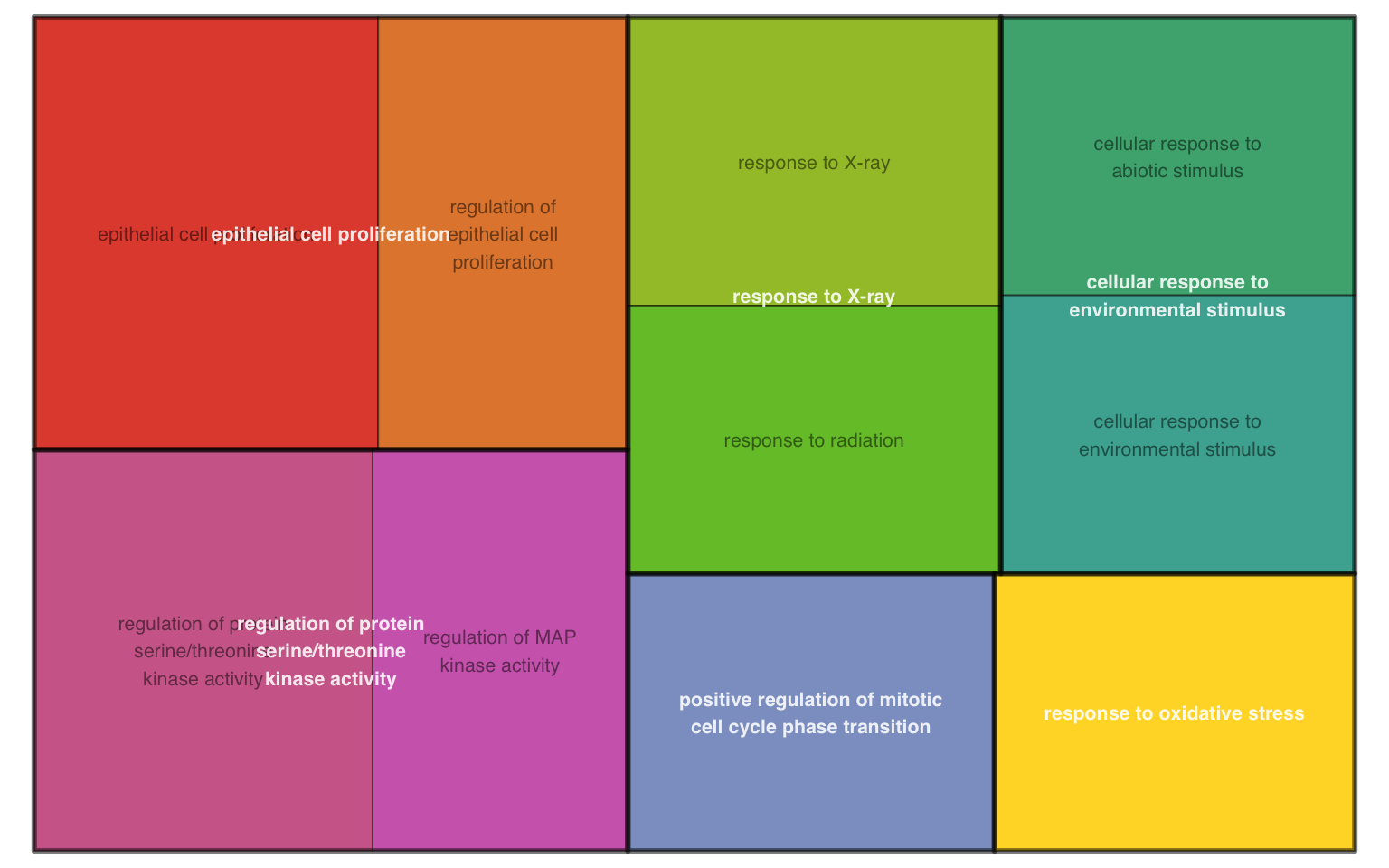
Figure 11.14: GO tangram plot of enrichment analysis.
11.15 Plot Theme
Genekitr provides a function plot_theme() to modify all plots theme including text size, border, legend, color, etc.
library(patchwork)
p1 <- plotEnrich(ego, plot_type = "dot")
p2 <- plotEnrich(ego,
plot_type = "dot",
main_text_size = 10,
legend_text_size = 10
)
p3 <- plotEnrich(ego,
plot_type = "dot",
border_thick = 3,
remove_grid = F
)
p4 <- plotEnrich(ego,
plot_type = "dot",
remove_main_text = T,
remove_legend_text = T,
remove_legend = T
)
p1 + p2 + p3 + p4 + plot_annotation(tag_levels = "A")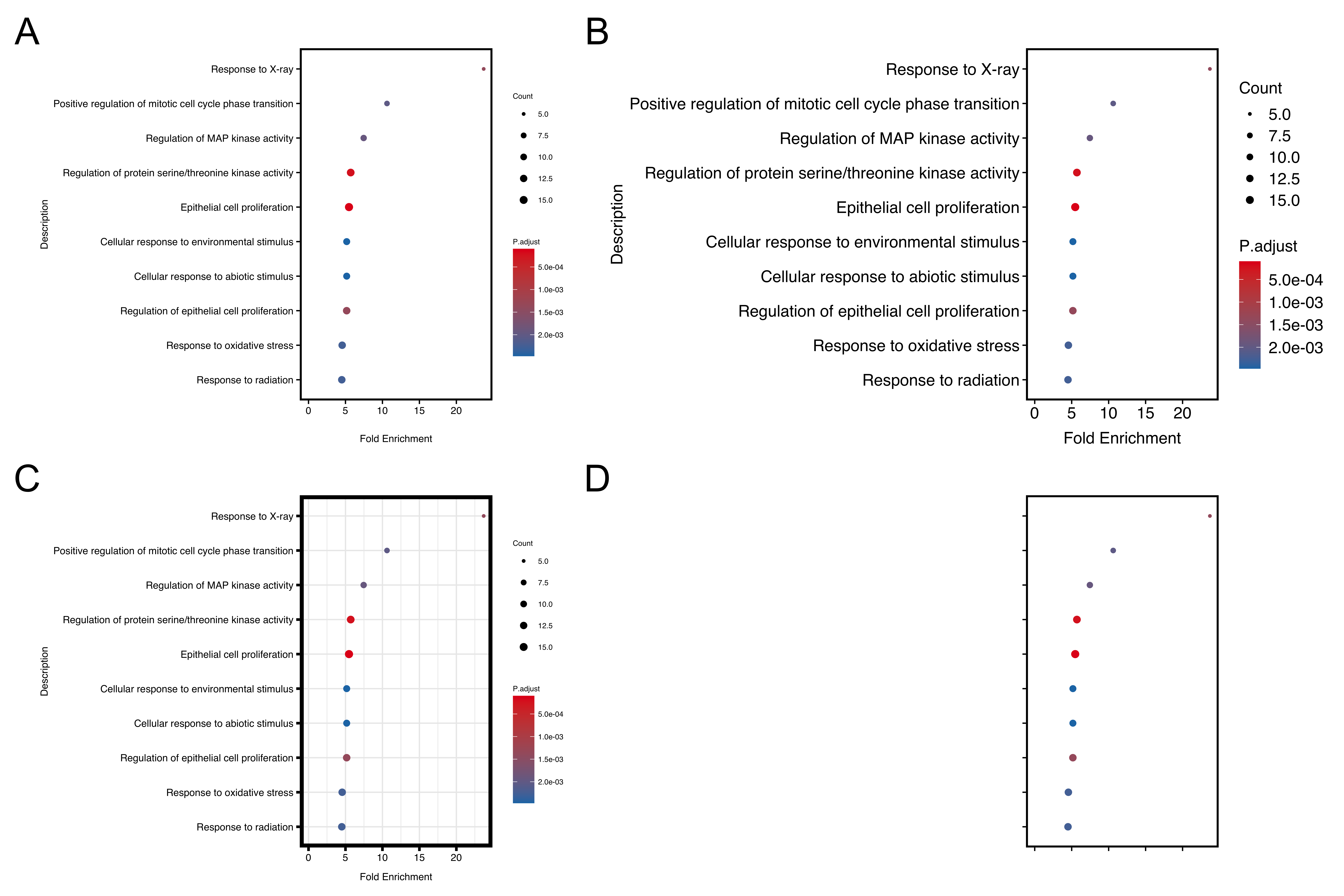
Figure 11.15: Plot theme. default theme (A), modify text size (B), modify grid line and border size (C) and remove all text and legend (D).
11.16 Advanced Plot
11.16.1 Two-group barplot for up/down regulated pathways
Up and down regulated genes could pass to genORA separately and visualize together as two-group barplot.
Here we take GO result as an example:
# 1st step: prepare input IDs
# Since the geneList is logFC decreasing ordered, we could take first 100 as up-regulated genes and vice versa.
data(geneList, package = "genekitr")
up_genes <- head(names(geneList), 100)
down_genes <- tail(names(geneList), 100)
# 2nd step: prepare gene set
hg_gs <- geneset::getGO(org = "human",ont = "bp")
# 3rd step: ORA analysis separately
up_go <- genORA(up_genes, geneset = hg_gs)
down_go <- genORA(down_genes, geneset = hg_gs)
dim(up_go)## [1] 50 12dim(down_go)## [1] 253 12There are two visulization types:
-
plot_type = "one": both up and down-regulated pathways are plotted together -
plot_type = "two": up and down-regulated pathways are plotted separately. Recommended if the number of both groups is similar.
plotEnrichAdv(up_go, down_go,
plot_type = "one",
term_metric = "FoldEnrich",
stats_metric = "p.adjust",
xlim_left = 25, xlim_right = 15) +
theme(legend.position = c(0.15, 0.9))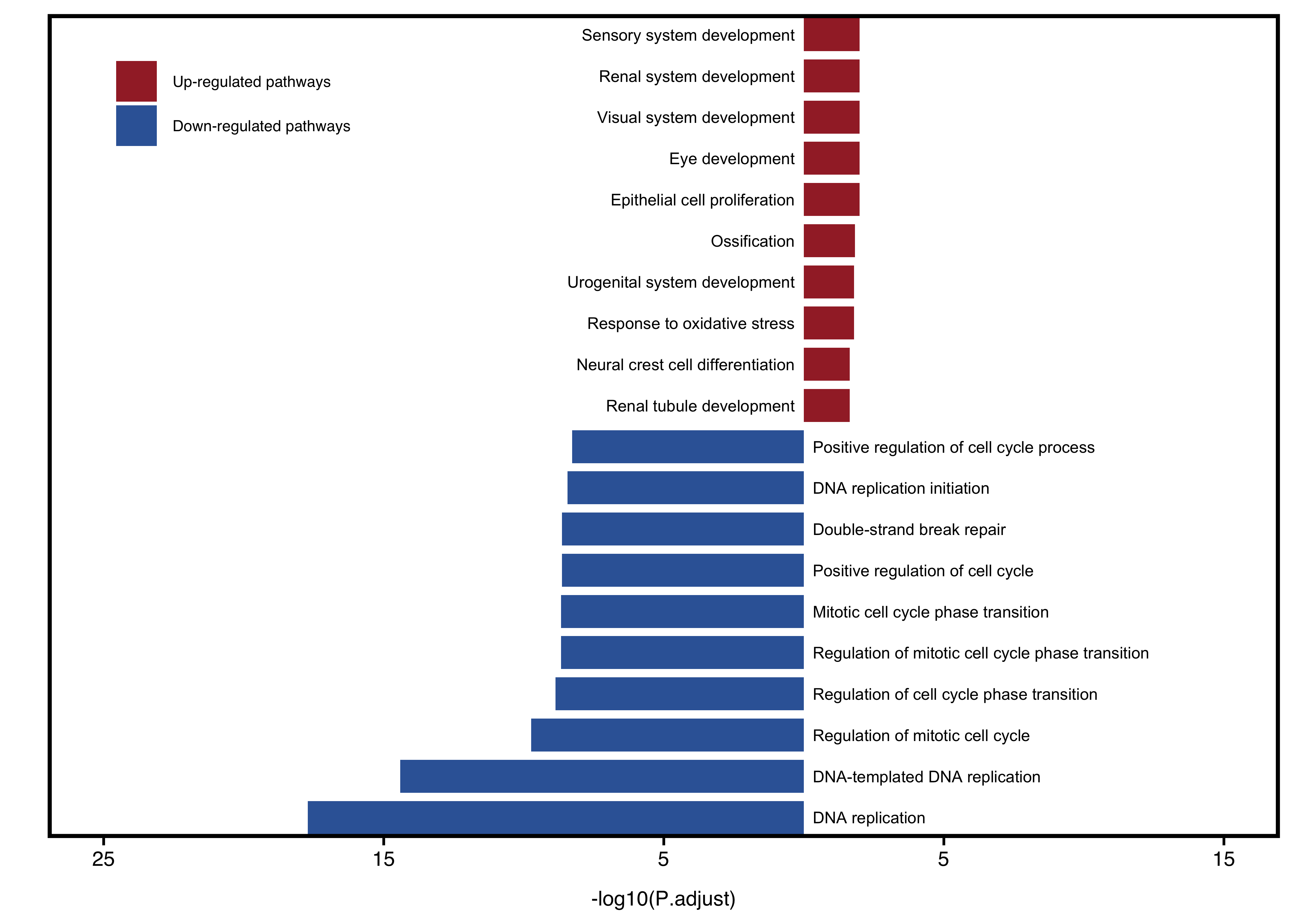
Figure 11.16: Visualize two groups together when plot_type = "one".
plotEnrichAdv(up_go, down_go,
plot_type = "two",
term_metric = "FoldEnrich",
stats_metric = "qvalue"
)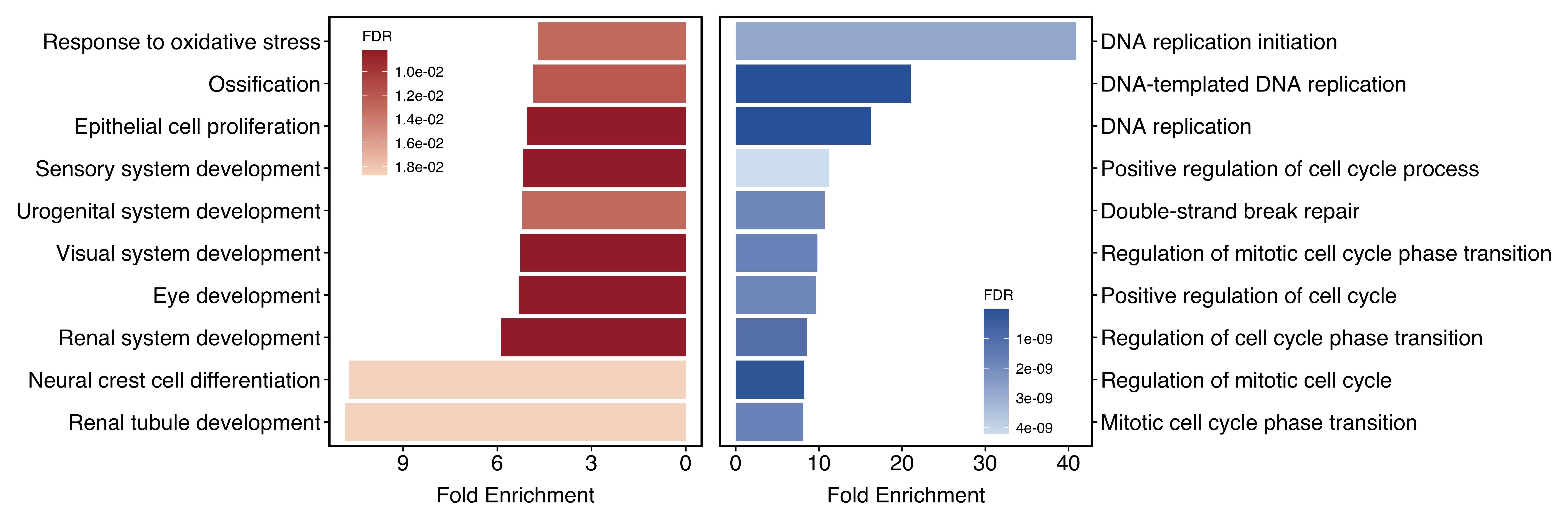
Figure 11.17: Visualize two groups seperately when plot_type = "two".